As stated in Lab 2, microorganisms exist in nature as mixed populations. However, to study microorganisms in the laboratory we must have them in the form of a pure culture, that is, one in which all organisms are descendants of the same organism.
Two major steps are involved in obtaining pure cultures from a mixed population:
1. First, the mixture must be diluted until the various individual microorganisms become separated far enough apart on an agar surface that after incubation, they form visible colonies isolated from the colonies of other microorganisms. This plate is called an isolation plate.
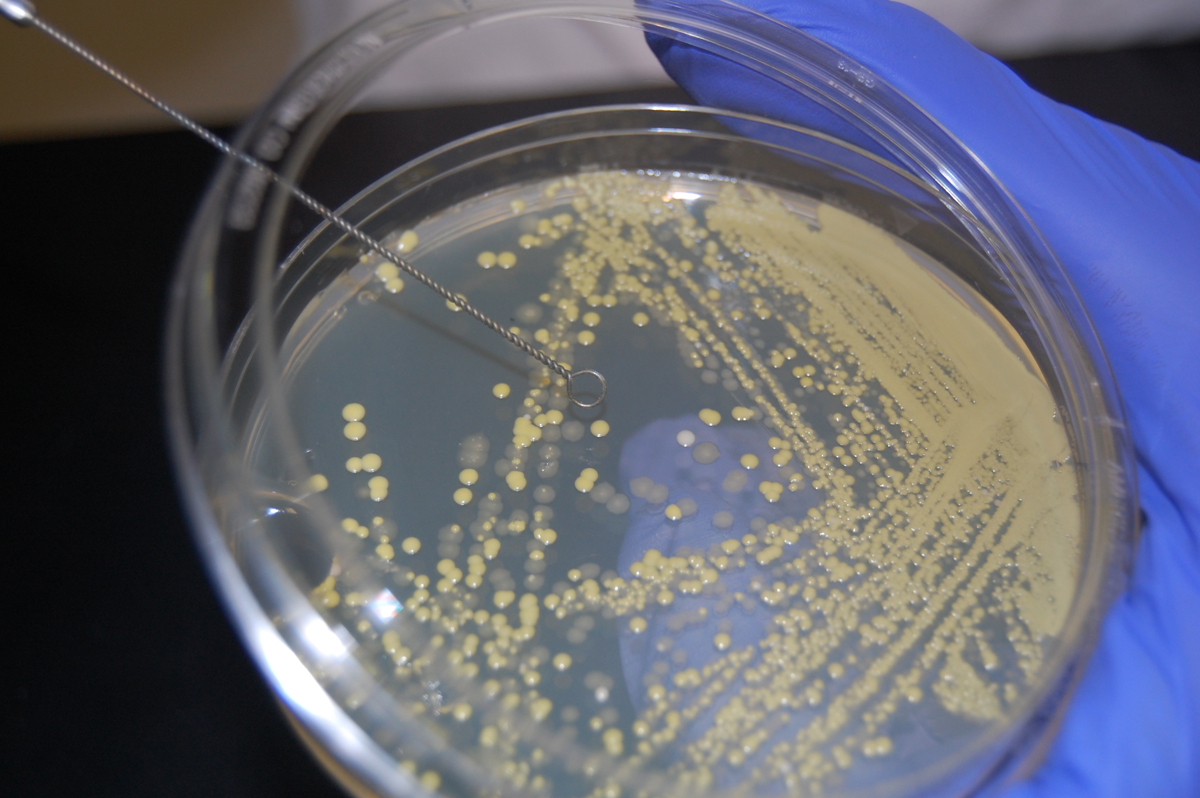
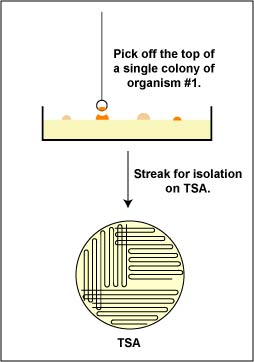
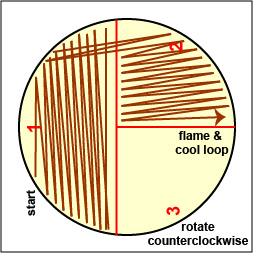
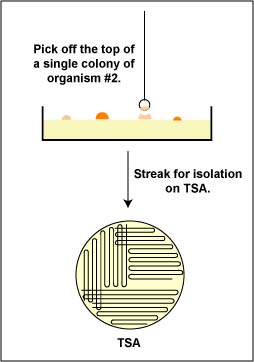
2. Then, an isolated colony can be aseptically "picked off" the isolation plate. (See Fig. 1.) and transferred to new sterile medium. (See Fig. 2.). After incubation, all organisms in the new culture will be descendants of the same organism, that is, a pure culture.
Fig. 1: Picking a Single Colony 0ff of a Petri Plate in order to Obtain a Pure Culture Fig. 2: Obtaining Pure Cultures from an Isolation Plate
Before removing bacteria from the petri plate, first cool the loop by sticking it into the agar away from any growth.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
A. STREAK PLATE METHOD OF ISOLATION
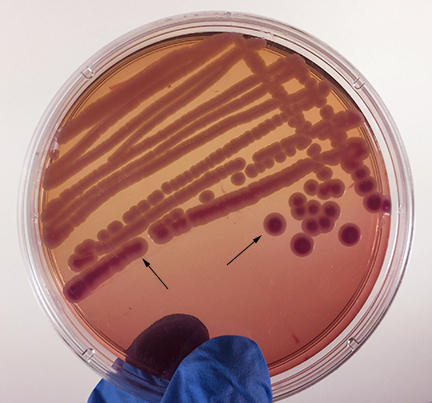
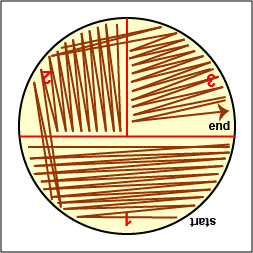
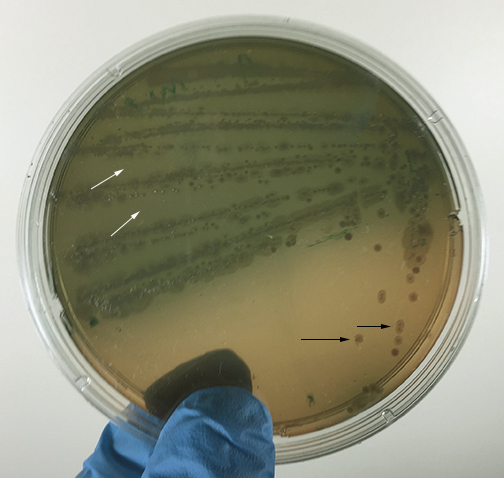
The most common way of separating bacterial cells on the agar surface to obtain isolated colonies is the streak plate method we used in Lab 2 to inoculate a petri plate. It provides a simple and rapid method of diluting the sample by mechanical means. As the loop is streaked across the agar surface, more and more bacteria are rubbed off until individual separated organisms are deposited on the agar. After incubation, the area at the beginning of the streak pattern will show confluent growth while the area near the end of the pattern should show discrete colonies. (See Fig. 3A and Fig. 3B.).
Fig. 3A: Isolation Plate: Mixture of Escherichia coli and Micrococcus luteus Grown on TSA
Fig. 3B: Isolation Plate: Mixture of Escherichia coli and Staphylococcus epidermidis Grown on TSA
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
| Aseptic Technique: Inoculation of broth tubes, slant tubes, and stab tubes |
| Aseptic Technique: Inoculating a Petri plate: Streaking for Isolation |
B. THE POUR PLATE AND SPIN PLATE METHODS OF ISOLATION
Another method of separating bacteria is the pour plate method. With the pour plate method, the bacteria are mixed with melted agar until evenly distributed and separated throughout the liquid. The melted agar is then poured into an empty plate and allowed to solidify. After incubation, discrete bacterial colonies can then be found growing both on the agar and in the agar.
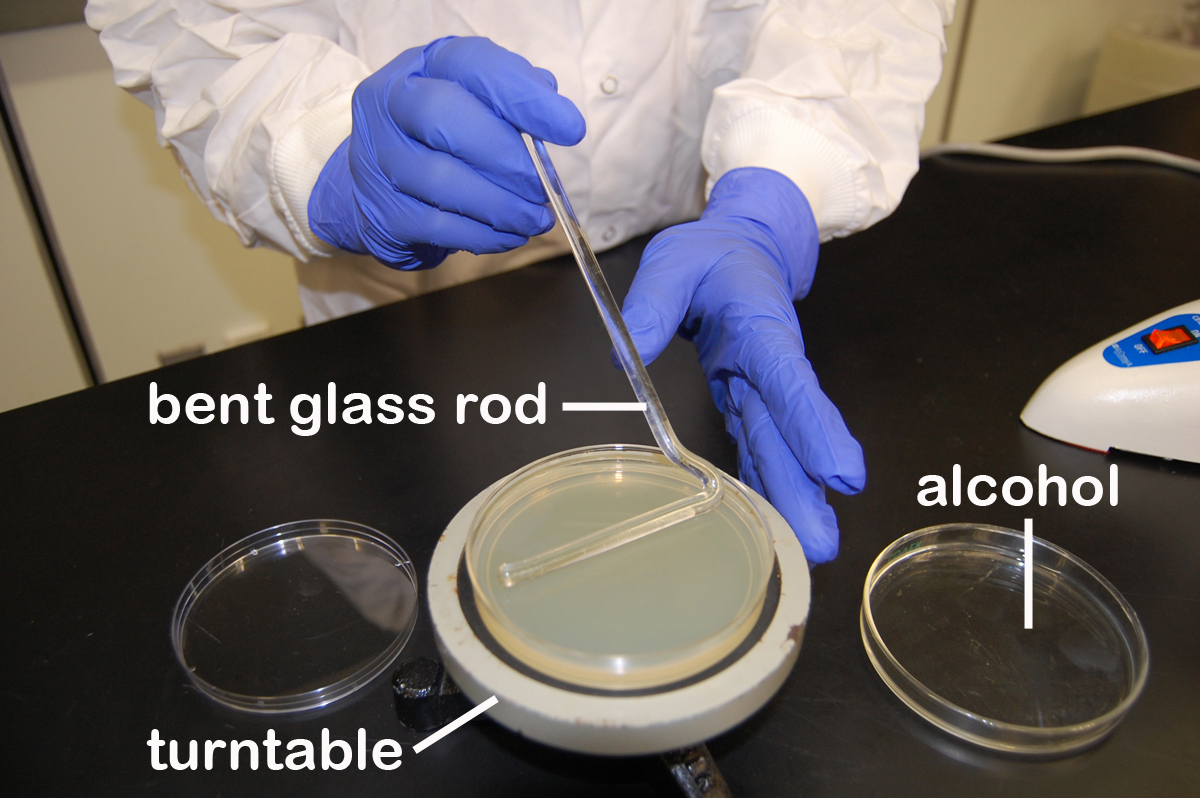
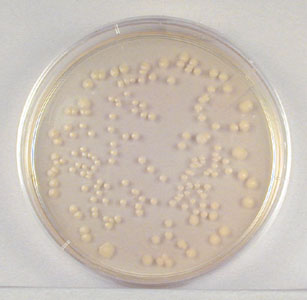
The spin plate method involves diluting the bacterial sample in tubes of sterile water, saline, or broth. Small samples of the diluted bacteria are then pipetted onto the surface of agar plates. A sterile, bent-glass rod is then used to spread the bacteria evenly over the entire agar surface. (See Fig. 4.) in order to see isolated colonies. (See Fig. 5.) In Lab 4 we will use this technique as part of the plate count method of enumerating bacteria.
Fig. 4: Using a Bent Glass Rod and a Turntable to Spread a Bacterial Sample
Fig. 5:Single Colonies from the Spin-Plate Method
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
C. USE OF SPECIALIZED MEDIA
To supplement mechanical techniques of isolation such as the streak plate method, many special-purpose media are available to the microbiologist to aid in the isolation and identification of specific microorganisms. These special purpose media fall into four groups: selective media, differential media, enrichment media, and combination selective and differential media.
1. Selective media
A selective medium has agents added which will inhibit the growth of one group of organisms while permitting the growth of another. For example, Columbia CNA agar has the antibiotics colistin and nalidixic acid added which inhibit the growth of Gram-negative bacteria but not the growth of Gram-positives. It is, therefore, said to be selective for Gram-positive organisms, and would be useful in separating a mixture of Gram-positive and Gram-negative bacteria.
Remember that even if you can only see the Gram-positive bacterium growing on a plate of Columbia CNA agar that was streaked with a mixture of a Gram-positive and a Gram-negative bacterium, this is NOT a pure culture. The Gram-negative bacteria are inhibited, not killed, and they are still alive on the agar surface. They have just not grown sufficiently that colonies are visible to the naked eye. Colonies still must be picked off of the Columbia CNA agar and streaked on a new agar plate before you have obtained a pure culture.
2. Differential media
A differential medium contains additives that cause an observable color change in the medium when a particular chemical reaction occurs. They are useful in differentiating bacteria according to some biochemical characteristic. In other words, they indicate whether a certain organism can carry out a specific biochemical reaction during its normal metabolism. Many such media will be used in future labs to aid in the identification of microorganisms.
3. Enrichment media
An enrichment medium contains additives that enhance the growth of certain organisms. This is useful when the organism you wish to culture is present in relatively small numbers compared to the other organisms growing in the mixture.
4. Combination selective and differential media
A combination selective and differential medium permits the growth of one group of organisms while inhibiting the growth of another. In addition, it differentiates those organisms that grow based on whether they can carry out particular chemical reactions.
For example, MacConkey agar. (See Fig. 6A.) is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives. If the Gram-negative bacterium ferments the sugar lactose in the medium, the acid end products lower the pH of the medium. The neutral red in the agar turns red in color once the pH drops below 6.8. As the pH drops, the neutral red is absorbed by the bacteria, causing the colonies to appear bright pink to red.
Results are interpreted as follows:
- Strong fermentation of lactose with high levels of acid production by the bacteria causes the colonies and confluent growth to appear bright pink to red. The resulting acid, at high enough concentrations, can also causes the bile salts in the medium to precipitate out of solution causing a pink precipitate (cloudiness) to appear in the agar surrounding the growth .(See Fig. 6B.)
- Weak fermentation of lactose by the bacteria causes the colonies and confluent growth to appear pink to red, but without the precipitation of bile salts there is no pink precipitate (cloudiness) in the agar surrounding the growth .(See Fig. 6C.)
- If the bacteria do not ferment lactose, the colonies and confluent growth appear colorless and the agar surrounding the bacteria remains relatively transparent. (See Fig. 6D.)
Fig. 6A: Uninoculated Plate of MacConkey Agar
Fig. 6B: Escherichia coli Growing on MacConkey Agar
Fig. 6C: Klebsiella aerogenes (formerly known as Enterobacter aerogenes) Growing on MacConkey Agar
Fig. 6D: Proteus mirabilis Growing on MacConkey Agar
MacConkey agar is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives.
MacConkey agar is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives.Strong fementation of lactose with high levels of acid production by the bacteria causes the colonies and confluent growth to appear bright pink to red. The resulting acid, at high enough concentrations, can also causes the bile salts in the medium to precipitate out of solution causing a pink precipitate (cloudiness) o appear around the the growth (arrows).
MacConkey agar is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives. Weak fermentation of lactose by the bacteria causes the colonies and confluent growth to appear pink or pink with red centers (arrows), but without the precipitation of bile salts there is no pink halo around the growth.
MacConkey agar is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives.If the bacteria do not ferment lactose, the colonies and confluent growth appear colorless and the agar surrounding the bacteria remains relatively transparent (arrow).
Photograph from From MicrobeLibrary.org
Courtesy of David Miller and Patrick Hanley, Hartwick College, Oneonta, NY.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Typical colony morphology on MacConkey agar is as follows:
Escherichia coli: colonies and confluent growth appear bright pink to red and surrounded by a pink precipitate (cloudiness) in the agar surrounding the growth. (See Fig. 6B.)
Enterobacter and Klebsiella: colonies and confluent growth appear bright pink to red but are not surrounded by a pink precipitate (cloudiness) in the agar surrounding the growth. (See Fig. 6C.)
Pseudomonas, Salmonella, Serratia, Proteus, and Shigella: colorless colonies; agar relatively transparent. (See Fig. 6C.)
Remember that even if you can only see the the Gram-negative bacterium growing on a plate of MacConkey agar that was streaked with a mixture of a Gram-positive and a Gram-negative bacterium, this is NOT a pure culture. The Gram-positive bacteria are inhibited, not killed, and they are still alive on the agar surface. They have just not grown sufficiently that colonies are visible to the naked eye. Colonies still have to be picked off of the MacConkey agar and streaked on a new agar plate before you have obtained a pure culture.
There are literally hundreds of special-purpose media available to the microbiologist. Today we will combine both a mechanical isolation technique (the streak plate) with selective and selective-differential media to obtain pure cultures from a mixture of bacteria. In future labs, such as 12 - 16, which deal with the isolation and identification of pathogenic bacteria, we will use many additional special-purpose media.
MEDIA
One plate of each of the following media: Trypticase Soy agar, Columbia CNA agar, and MacConkey agar.
ORGANISMS
A broth culture containing a mixture of one of the following Gram-positive bacteria and one of the following Gram-negative bacteria:
- Possible Gram-positive bacteria:
- Micrococcus luteus.
A Gram-positive coccus with a tetrad or a sarcina arrangement; produces
circular, convex colonies with a yellow, water-insoluble pigment on Trypticase
Soy agar and Columbia CNA agar.
- Micrococcus luteus growing on TSA.(See Fig. 7A)
- Close up ofMicrococcus luteus growing on TSA. (See Fig. 7B.)
- Staphylococcus epidermidis.
A Gram-positive coccus with a staphylococcus arrangement; produces circular,
convex, non-pigmented colonies on Trypticase Soy agar and Columbia CNA agar.
- Staphylococcus epidermidis growing on TSA. (See Fig. 7C)
- Close up of Staphylococcus epidermidis growing on TSA. (See Fig. 7D.)
Fig. 7A: Isolation Plate: Mixture of Escherichia coli and Micrococcus luteus Grown on TSA
Fig. 7B: Close Up of Escherichia coli and Micrococcus luteus Grown on TSA
Fig. 7C: Isolation Plate: Mixture of Escherichia coli and Staphylococcus epidermidis Grown on TSA
Fig. 7D: Close Up of Escherichia coli and Staphylococcus epidermidis Grown on TSA
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

- Micrococcus luteus.
A Gram-positive coccus with a tetrad or a sarcina arrangement; produces
circular, convex colonies with a yellow, water-insoluble pigment on Trypticase
Soy agar and Columbia CNA agar.
- Possible Gram-negative bacteria:
- Escherichia coli.
A Gram-negative bacillus; produces irregular, raised, non-pigmented colonies on Trypticase Soy agar.
- Escherichia coli growing on TSA. (See Fig. 7E.)
- Klebsiella aerogenes.
A Gram-negative bacillus; produces irregular raised, non-pigmented colonies on Trypticase Soy agar.
- Klebsiella aerogenes (formerly known as Enterobacter aerogenes) growing on TSA. (See Fig. 7F.)
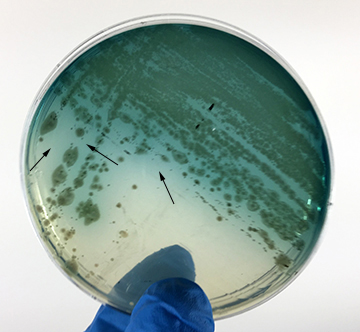
- Pseudomonas aeruginosa. A Gram-negative bacillus; produces irregular raised colonies and a green to blue water-soluble pigment on Trypticase soy agar.
- Pseudomonas aeruginosa growing on TSA. (See Fig. 7G.)
- Escherichia coli.
A Gram-negative bacillus; produces irregular, raised, non-pigmented colonies on Trypticase Soy agar.
Fig. 7E: Isolation Plate: Mixture of Escherichia coli and Micrococcus luteus Grown on TSA
Fig. 7F: Mixture of Klebsiella aerogenes (formerly known as Enterobacter aerogenes) and Staphylococcus epidermidis Grown on TSA
Fig. 7G: Pseudomonas aeruginosa Growing on TSA
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
During the next two labs you will attempt to obtain pure cultures of each organism in your mixture and determine which two bacteria you have. Today you will try to separate the bacteria in the mixture in order to obtain isolated colonies; next lab you will identify the two bacteria in your mixture and pick off single isolated colonies of each of the two bacteria in order to get a pure culture of each. The following lab you will prepare microscopy slides of each of the two pure cultures to determine if they are indeed pure.
PROCEDURE (to be done in pairs)
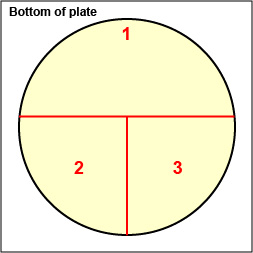
1. On the bottom of each of the 3 petri plate you are using today, divide the plate into thirds with your wax marker and label as shown in Fig. 8A below. This will guide your streaking.
2. Although Trypticase Soy agar (TSA), which grows both Gram-positive and Gram-negative bacteria, is not normally used as an isolation medium, we will attempt to obtain isolated colonies of the two organisms in your mixture by using strictly mechanical methods. Often, however, one bacterium overgrows another in a mixture and by the time you spread out the more abundant organism enough to get isolated colonies, the one in smaller numbers is no longer on the loop so you may not see single colonies of each on the TSA next time.
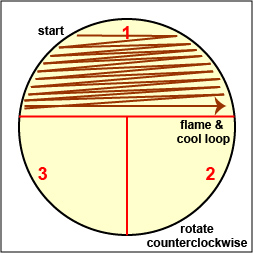
Streak your mixture on a plate of Trypticase Soy agar as illustrated in Figs. 8B, 8C, and 8D.
Fig. 8A: Label Plate to Guide your Streaking Fig. 8B: Streaking For Isolation, Step-1
Fig. 8C: Streaking For Isolation, Step-2
Fig. 8D: Streaking For Isolation, Step-3
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
3. Streak the same mixture for isolation on a plate of Columbia CNA agar as illustrated in Figs. 8B, 8C, and 8D. Columbia CNA agar is selective for Gram-positive bacteria.
- Micrococcus luteus growing on Columbia CNA agar. See Fig. 8E.
- Staphylococcus epidermidis growing on Columbia CNA agar. See Fig. 8F.
Fig. 8E: Micrococcus luteus Growing on Columbia CNA Agar
Staphylococcus epidermidis> Growing on Columbia CNA Agar
Only Gram-positive bacteria grow on Columbia CNA agar. (Gram-negative bacteria are inhibited by the colistin and nalidixic acid added to the agar.) Micrococcus luteus produces a yellow, water insoluble pigment.
Only Gram-positive bacteria grow on Columbia CNA agar. (Gram-negative bacteria are inhibited by the colistin and nalidixic acid added to the agar.) Staphylococcus epidermidis does not normally produce pigment.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
4. Streak the same mixture for isolation on a plate of MacConkey agar as illustrated in Figs. 8B, 8C, and 8D above. MacConkey agar is selective for Gram-negative bacteria and differential for certain members of the bacterial family Enterobacteriaceae as well as Pseudomonas.
- Escherichia coli growing on MacConkey agar. See Fig. 8G.
- Klebsiella aerogenes (formerly known as Enterobacter aerogenes) growing on MacConkey agar. See Fig. 8H.
- Pseudomonas aeruginosa growing on MacConkey agar. See Fig. 8I.
Fig. 8G: Escherichia coli Growing on MacConkey Agar
Fig. 8H: Klebsiella aerogenes (formerly known as Enterobacter aerogenes) Growing on MacConkey Agar
Fig. 8I: Pseudomonas aeruginosa Growing on MacConkey Agar
Strong fementation of lactose with high levels of acid production by the bacteria causes the colonies and confluent growth to appear bright pink to red. The resulting acid, at high enough concentrations, can also causes the bile salts in the medium to precipitate out of solution causing a pink precipitate (cloudiness) o appear around the the growth (arrows).
Weak fermentation of lactose by the bacteria causes the colonies and confluent growth to appear pink or pink with red centers (arrows), but without the precipitation of bile salts there is no pink halo around the growth.
If the bacteria do not ferment lactose, the colonies and confluent growth appear relatively colorless and the agar surrounding the bacteria remains relatively transparent. The green, water-soluble pigment produced by Pseudomonas aeruginosa (white arrows) is also evident here.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
5. Incubate the three plates upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section until the next lab period.
RESULTS
1. Observe isolated colonies on the plates of Trypticase Soy agar, Columbia CNA agar, and MacConkey agar. Record your observations and conclusions.
2. Using any of the three plates on which they are growing:
a. Aseptically pick off a single isolated colony of each of the two bacteria from your original mixture that you have just identified and aseptically transfer them to separate plates of Trypticase Soy agar. (See Fig. 9). Remember to streak the plate for isolation as shown in Figs. 8B, 8C, and 8D above.
Fig. 9: Obtaining Pure Cultures from an Isolation Plate
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
b. When picking off single colonies, remove the top portion of the colony without touching the agar surface itself to avoid picking up any inhibited bacteria from the surface of the agar. Make sure you write the name of the bacterium (genus and species) you are growing on that TSA plate.
c. Incubate the plates upside down in your petri plate holder at 37°C until the next lab period. These will be your pure cultures for Lab 5 (Direct and Indirect stains).
PERFORMANCE OBJECTIVES FOR LAB 3
After completing this lab, the student will be able to complete the following objectives:
DISCUSSION
1. Given a mixture of a Gram-positive and a Gram-negative bacterium and plates of Columbia CNA, MacConkey, and Trypticase Soy agar, describe the steps you would take to eventually obtain pure cultures of each organism.
2. Define: selective medium, differential medium, enrichment medium, and combination selective-differential medium.
3. State the usefulness of Columbia CNA agar and MacConkey agar.
4. Describe how each of the following would appear when grown on MacConkey agar:
a. Escherichia coli
b. Klebsiella aerogenes (formerly known as Enterobacter aerogenes)
c. Pseudomonas aeruginosaPROCEDURE
1. Using the streak plate method of isolation, obtain isolated colonies from a mixture of microorganisms.
2. Pick off isolated colonies of microorganisms growing on a streak plate and aseptically transfer them to sterile media to obtain pure cultures.
RESULTS
1. When given a plate of Columbia CNA agar or MacConkey agar showing discrete colonies, correctly interpret the results.
SELF-QUIZ
Microbiology Laboratory Manual by Gary E. Kaiser, PhD, Professor of Microbiology
is licensed under a Creative Commons Attribution 4.0 International License.
Last updated: March, 2023
Please send comments and inquiries to Dr. Gary Kaiser