INTRODUCTION
DISCUSSION
In the three previous labs we examined bacteria microscopically. Staining provides valuable information as to bacterial morphology, gram reaction, and presence of such structures as capsules and endospores. Beyond that, however, microscopic observation gives little additional information as to the genus and species of a particular bacterium.
To identify bacteria, we must rely heavily on biochemical testing. The types of biochemical reactions each organism undergoes act as a "thumbprint" for its identification. This is based on the following chain of logic:
- Each different species
of bacterium has a different molecule of DNA (i.e., DNA with
a unique series of nucleotide bases).
- Since DNA codes for protein
synthesis, then different species of bacteria must, by way of their
unique DNA, be able to synthesize different protein enzymes.
- Enzymes catalyze all
the various chemical reactions of which the organism is capable. This
in turn means that different species of bacteria must carry out different
and unique sets of biochemical reactions.
Lab 7 will demonstrate that different bacteria, because of their unique enzymes, are capable of different biochemical reactions. It will also show the results of the activity of those enzymes. In later labs we will use a wide variety of special purpose differential media frequently used in the clinical laboratory to identify specific pathogenic and opportunistic bacteria.
In general, we can classify enzymes as being either exoenzymes or endoenzymes. Exoenzymes are secreted by bacteria into the surrounding environment in order to break down larger nutrient molecules so they may enter the bacterium . Once inside the organism, some of the nutrients are further broken down to yield energy for driving various cellular functions, while others are used to form building blocks for the synthesis of cellular components. These later reactions are catalyzed by endoenzymes located within the bacterium.
- If the bacterium ferments that particular carbohydrate producing acid end products alone, the acid will lower the pH, causing the pH indicator phenol red to change form its original red color at a neutral pH to a yellow or clear color. See Fig. 8A.
- If the bacterium ferments that particular carbohydrate producing both acid and gas, the pH indicator phenol red to change form its original red color at a neutral pH to a yellow or clear color and the gas will collect in the Durham tube as a substantial gas bubble appearing at the top of the Durham tube. See Fig. 8B.
- If the bacterium does not ferment the carbohydrate, no acid or gas will be produced. The phenol red will remain red and there will be no gas bubble seen at the top of the Durham tube. See Fig. 8C.
- If the bacterium produced an exoenzyme that hydrolized the starch in the agar, a clear zone will surrround the bacterial growth because the starch is no longer there to react with the iodine (see Fig. 11A).
- If the bacterium lacks the exoenzyme to break down the starch, the agar around the growth should turn dark brown or blue/black color (see Fig. 11B) due to the iodine-starch complex.
- If the bacterium produced an exoenzyme capable of hydrolyzing the casein, there will be a clear zone around the bacterial growth (see Fig. 12A).
- If the bacterium lacks the exoenzyme to break down casein, the Skim Milk agar will remain white and opaque (see Fig. 12B).
- A
change in color in the tube from red or orange to yellow or clear indicates
that the organism has fermented that carbohydrate, producing
acid end products.
- A substantial gas
bubble at the top of the Durham tube, the inverted test tube within
the broth, indicates
gas was also produced from the fermentation of the
carbohydrate.
- If the phenol red remains red, no acid was produced and the carbohydrate was not fermented.
- Carbohydrate fermentation producing
acid but no gas: acidic (yellow or clear); no substantial
gas bubble in the Durham tube (see
Fig. 13A).
- Carbohydrate fermentation producing
acid and gas: acidic (yellow or clear) ; a substantial
gas bubble in the Durham tube (see
Fig. 13B).
- No carbohydrate fermentation.
No acid or gas (neutral pH (red or orange); no substantial
gas bubble
in the Durham tube (see
Fig. 13C).
- If the bacterium produces the enzyme to reduce sulfur to hydrogen sulfide (H2S), the agar will turn black indicating that the organism has produced hydrogen sulfide. See Fig. 14A.
- If the bacterium lacks the enzyme, the agar does not turn black, indicating that hydrogen sulfide was not produced.
- If the bacterium produces the enzyme to break down tryptophan into molecules of indole, pyruvic acid, and ammonia, the Kovac's reagent will turn red, indicating the organism is indole positive. See Fig. 14B.
- If the Kovac's reagent remains yellow, no indole was produced and the organism is indole negative. See Fig. 14C.
- If the bacterium produces the enzyme catalase, then the hydrogen peroxide added to the culture will be broken down into water and free oxygen. The oxygen will bubble through the water causing a surface froth to form. This is a catalase positive bacterium (see fig.15A).
- A catalase negative bacterium will not produce catalase to break down the hydrogen peroxide, and no frothing will occur (see Fig. 15B).
| Aseptic Technique Tips |
| Aseptic Technique: Inoculation of broth tubes, slant tubes, and stab tubes |
A. STARCH HYDROLYSIS
DISCUSSION
Starch is a polysaccharide which appears as a branched polymer of the simple sugar glucose. This means that starch is really a series of glucose molecules hooked together to form a long chain. Additional glucose molecules then branch off of this chain as shown below.
|
( ---GLU-GLU-GLU-GLU-GLU-GLU-GLU--- )n
Some bacteria are capable of using starch as a source of carbohydrate but in order to do this, they must first hydrolyze or break down the starch so it may enter the cell. The bacterium secretes an exoenzyme which hydrolyzes the starch by breaking the bonds between the glucose molecules. This enzyme is called a diastase.
action of diastase
The glucose can then enter the bacterium and be used for metabolism.
MEDIUM
Starch agar (one plate) See Fig. 1.
ORGANISMS
Trypticase Soy broth cultures of Bacillus subtilis and Escherichia coli.
PROCEDURE (to be done in pairs)
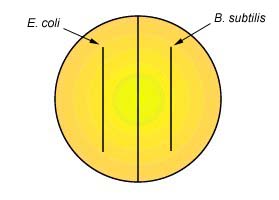
1. Using a wax marker, draw a line on the bottom of a Starch agar plate so as to divide the plate in half. Label one half B. subtilis and the other half E. coli.
2. Make a single streak line with the appropriate organism on the corresponding half of the plate as shown in Fig. 2.
3. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section until the next lab period.
4. Next period, iodine will be added to see if the starch remains in the agar or has been hydrolyzed by the exoenzyme diastase. Iodine reacts with starch to produce a dark brown or blue/black color. If starch has been hydrolyzed there will be a clear zone around the bacterial growth (see Fig. 3A) because the starch is no longer in the agar to react with the iodine. If starch has not been hydrolyzed, the agar will remain a dark brown or blue/black color (see Fig. 3B).
Fig. 1: Starch Agar Fig. 2: Inoculation of Starch Agar
Fig. 3A: Starch Hydrolysis by Bacillus subtilis on Starch Agar
Fig. 3B: No Starch Hydrolysis by Escherichia coli on Starch Agar
Starch agar before inoculation.
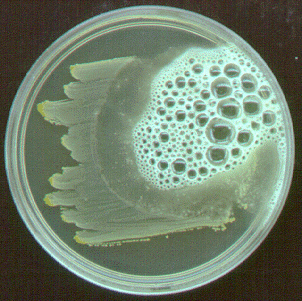
The clear zone around the growth of Bacillus subtilis no longer contains starch to react with the iodine. The starch was hydrolized by a diastase produced by the bacterium.
The starch was not hydrolized by the Escherichia coli on the left. The starch reacts with the iodine producing the dark color.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
B. PROTEIN HYDROLYSIS
DISCUSSION
Proteins are made up of various amino acids linked together in long chains by means of peptide bonds. Many bacteria can hydrolyze a variety of proteins into peptides (short chains of amino acids) and eventually into individual amino acids. They can then use these amino acids to synthesize their own proteins and other cellular molecules or to obtain energy. The hydrolysis of protein is termed proteolysis and the enzyme involved is called a protease. In this exercise we will test for bacterial hydrolysis of the protein casein, the protein that gives milk its white, opaque appearance.
MEDIUM
Skim Milk agar (one plate) See Fig. 4.
ORGANISMS
Trypticase Soy broth cultures of Bacillus subtilis and Escherichia coli.
PROCEDURE (to be done in pairs)
1. Divide the Skim Milk agar plate in half and inoculate one half with Bacillus subtilis and the other half with Escherichia coli as done above with the above starch agar plate (see Fig. 5).
2. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section until the next lab period. If casein is hydrolyzed, there will be a clear zone around the bacterial growth (see Fig 6A). If casein is not hydrolyzed, the agar will remain white and opaque (see Fig. 6B).
Fig. 4: Skim Milk Agar
Fig. 5: Inoculation of Skim Milk Agar
Fig. 6A: Casein Hydrolysis by Bacillus subtilis on Skim Milk Agar
Fig. 6B: No Casein Hydrolysis by Escherichia coli on Skim Milk Agar
Skim milk agar before inoculation.
The clear zone around the growth of the Bacillus subtilis on the right no longer contains casein. The casein was hydrolized by a protease produced by the bacterium.
The casein was not hydrolized by the Escherichia coli on the left. The agar remains white and opaque.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
C. FERMENTATION OF CARBOHYDRATES
DISCUSSION
Carbohydrates are complex chemical substrates which serve as energy sources when broken down by bacteria and other cells. They are composed of carbon, hydrogen, and oxygen (with hydrogen and oxygen being in the same ratio as water; [CH2O]) and are usually classed as either sugars or starches.
Facultative anaerobic and anaerobic bacteria are capable of fermentation, an anaerobic process during which carbohydrates are broken down for energy production. A wide variety of carbohydrates may be fermented by various bacteria in order to obtain energy and the types of carbohydrates which are fermented by a specific organism can serve as a diagnostic tool for the identification of that organism.
We can detect whether a specific carbohydrate is fermented by looking for common end products of fermentation. When carbohydrates are fermented as a result of bacterial enzymes, the following fermentation end products may be produced:
1. acid end products, or
2. acid and gas end products.
T`o test for these fermentation products, you inoculate and incubate tubes of media containing a single carbohydrate (such as lactose or maltose), a pH indicator (such as phenol red) and a Durham tube (a small inverted tube to detect gas production). See Fig. 7.
Fig. 7: An Uninoculated Fermentation Tube. Fig. 8A: A Fermentation Tube Showing Bacterial Fermentation of the Carbohydrate Producing Acid but No Gas Fig. 8B: A Fermentation Tube Showing Bacterial Fermentation of the Carbohydrate Producing both Acid and Gas. Fig. 8C: A Fermentation Tube Showing No Bacterial Fermentation of the Carbohydrate Producing No Acid or Gas.
Fermentation tube before inoculation. The phenol red remains red at a neutral pH and there is no gas bubble at the top of the Durham tube.
Acid end products from carbohydrate fermentation lower the pH causing the pH indicator phenol red to turn from red (neutral) to yellow (acid), but there is no gas bubble seen at the top of the Durham tube.
Acid end products from carbohydrate fermentation lower the pH causing the pH indicator phenol red to turn from red (neutral) to yellow (acid). A large bubble seen at the top of the Durham tube indicates gas end products.
Fermentation tube showing no carbohydrate fermentation. The phenol red remains red indicating no acid, and there is no bubble seen at the top of the Durham tube indicating no gas.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
MEDIA
3 tubes of Phenol Red Lactose broth and 3 tubes of Phenol Red Maltose broth.
ORGANISMS
Trypticase Soy agar cultures of Bacillus subtilis, Escherichia coli, and Staphylococcus aureus.
PROCEDURE (to be done in pairs)
1. Label each tube with the name of the sugar in the tube and the name of the bacterium you are growing.
2. Inoculate one Phenol Red Lactose broth tube and one Phenol Red Maltose broth tube with Bacillus subtilis.
3. Inoculate a second Phenol Red Lactose broth tube and a second Phenol Red Maltose broth tube with Escherichia coli.4. Inoculate a third Phenol Red Lactose broth tube and a third Phenol Red Maltose broth tube with Staphylococcus aureus.
5. Incubate the tubes in your test tube rack on your shelf of the 37°C incubator corresponding to your lab section until the next lab period
D. INDOLE AND HYDROGEN SULFIDE PRODUCTION
DISCUSSION
Sometimes we look for the production of products produced by only a few bacteria. As an example, some bacteria use the enzyme tryptophanase to convert the amino acid tryptophan into molecules of indole, pyruvic acid and ammonia. Since only a few bacteria contain tryptophanase, the formation of indole from a tryptophan substrate can be another useful diagnostic tool for the identification of an organism. Indole production is a key test for the identification of Escherichia coli.
Likewise, some bacteria are capable
of breaking down sulfur containing amino acids (cystine, methionine) or reducing
inorganic sulfur-containing compounds (such as sulfite, sulfate, or thiosulfate)
to produce hydrogen sulfide (H2S). This
reduced sulfur may then be incorporated into other cellular amino acids, or
perhaps into coenzymes. The ability of an organism to reduce sulfur-containing
compounds to hydrogen sulfide can be another test for identifying unknown organisms
such as certain Proteus and Salmonella. We can test for both indole production and hydrogen sulfide production using SIM medium. See Fig. 9A.
SIM medium contains tryptophan. By adding Kovac's reagent to the SIM medium after inoculation and incubation, we can determine if the bacteria produce indole. Kovac's reagent reacts with indole and turns red. See
Fig. 9B.
SIM medium also contains a sulfur-containing compound and iron salts. If the sulfur is reduced by the bacteria and hydrogen sulfide is produced, the hydrogen sulfide will combine with the iron salts to form visible black ferric sulfide (FeS) in the agar. See Fig. 9C.
If neither hydrogen sulfide nor indole are produced by the bacteria in the SIM medium, the agar does not turn black and the Kovac's reagent will remain yellow. See Fig. 9D.
Fig. 9A: Uninoculated SIM Medium
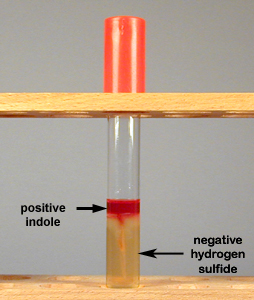
Fig. 9B: Production of Indole but No Hydrogen Sulfide in SIM Medium
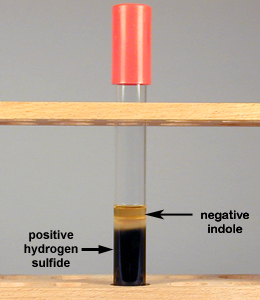
Fig. 9C: Production of Hydrogen Sulfide but No Indole in SIM Medium
Fig. 9D: Negative Hydrogen Sulfide and Negative Indole in SIM Medium
Sim medium before inoculation.
If indole is produced from the breakdown of the amino acid tryptophan, the Kovac's reagent, when added, will turn red. If hydrogen sulfide is not produced, the agar does not turn black.
SIM medium turns black if hydrogen sulfide is produced from the reduction of sulfur. If Kovac's reagent remains yellow, indole is not produced.
Indole negative, because the Kovac's reagent did not turn red, and hydrogen sulfide negative, because the agar did not turn black.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
MEDIUM
Three tubes of SIM (Sulfide, Indole, Motility) medium. This medium contains a sulfur source, an iron salt, the amino acid tryptophan, and is semi-solid in agar content (0.3%). It can be used to detect hydrogen sulfide production, indole production, and motility.
ORGANISMS
Trypticase Soy agar cultures of Proteus mirabilis, Escherichia coli, and Enterobacter cloacae.
PROCEDURE (to be done in pairs)
1. Stab one SIM medium tube with Proteus mirabilis.
2. Stab a second SIM medium tube with Escherichia coli.
3. Stab a third SIM medium tube with Enterobacter cloacae.
4 . Incubate the tubes in your test tube rack on your shelf of the 37°C incubator corresponding to your lab section until the next lab period
5. Next lab period add Kovac's reagent to each tube to detect indole production.
E. CATALASE ACTIVITY (Demonstration)
DISCUSSION
Catalase is the name of an enzyme found in most bacteria which initiates the breakdown of hydrogen peroxide (H2O2) into water (H2O) and free oxygen (O2).
During the normal process of aerobic respiration, hydrogen ions (H+)are given off and must be removed by the cell. The electron transport chain takes these hydrogen ions and combines them with half a molecule of oxygen (an oxygen atom) to form water (H2O). During the process, energy is given off and is trapped and stored in ATP. Water is then a harmless end product. Some cytochromes in the electron transport system, however, form toxic hydrogen peroxide (H2O2) instead of water and this must be removed. This is done by the enzyme catalase breaking the hydrogen peroxide into water and oxygen as shown above. Most bacteria are catalase-positive; however, certain genera that don't carry out aerobic respiration, such as the genera Streptococcus, Enterococcus, Lactobacillus, and Clostridium, are catalase-negative.
MATERIALS
Trypticase Soy agar cultures of Staphylococcus aureus and Streptococcus lactis, 3% hydrogen peroxide.
PROCEDURE (demonstration)
Add a few drops of 3% hydrogen peroxide to each culture and look for the release of oxygen as a result of hydrogen peroxide breakdown. This appears as foaming and indicates the organism is catalase-positive. See Fig. 10A. If the organism does not have the enzyme catalase, then the hydrogen peroxide will not be broken down into water and oxygen and no foaming will be seen. See Fig. 10B.
Fig. 10A: A positive Catalase Test.
Fig. 10B: A Negative Catalase Test.
A Catalase Test on Staphylococcus aureus, a catalase-positive bacterium.
A Catalase Test on Streptococcus lactis. The genus Streptococcus and the genus Enterococcus are catalase-negative.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
RESULTS
A. Starch Hydrolysis
| Video review - How to Interpret the Results of Starch Hydrolysis |
When iodine is added to starch, the iodine-starch complex that forms gives a characteristic dark brown or deep purple color reaction. If the starch has been hydrolyzed into glucose molecules by the diastase exoenzyme, it no longer gives this reaction.
Flood the surface of the Starch agar plate with gram's iodine.
Record your results and indicate which organism could hydrolyze the starch (+ = hydrolysis; - = no hydrolysis).
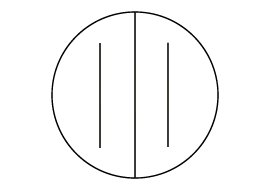
| Escherichia coli | Bacillus subtilis |
| Starch hydrolysis = | Starch hydrolysis = |
B. Protein Hydrolysis
| Video review - How to Interpret the Results of Casein Hydrolysis |
The protein casein exists as a colloidal suspension in milk and gives milk its characteristic white, opaque appearance. If the casein in the milk is hydrolyzed into peptides and amino acids, it will lose its opaqueness.
Record your results and indicate which organism could hydrolyze the casein (+ = hydrolysis; - = no hydrolysis).

| Escherichia coli | Bacillus subtilis |
| Casein hydrolysis = | Casein hydrolysis = |
C. Fermentation of Carbohydrates
| Video review - How to Interpret the Results of Carbohydrate Fermentation |
As mentioned above, we can detect whether a specific carbohydrate is fermented by looking for common end products of fermentation. When carbohydrates are fermented as a result of bacterial enzymes, the following fermentation end products may be produced:
1. acid end products, or
2. acid and gas end products.
The results of fermentation may be acid alone or acid plus gas, but never gas alone.
Phenol red pH indicator appears red or orange at neutral pH and appears yellow or clear at an acidic pH.
Possible results are as follows:
Record your results below (+ = positive; - = negative).
| Organism | Phenol Red Maltose | Phenol Red Lactose |
|---|---|---|
| Bacillus subtilis | ||
| Acid | ||
| Gas | ||
| Fermentation | ||
| Escherichia coli | ||
| Acid | ||
| Gas | ||
|
Fermentation
|
||
| Staphylococcus aureus | ||
| Acid | ||
| Gas | ||
| Fermentation |
D. Production of Indole and Hydrogen Sulfide
| Video review - How to Interpret the Results of SIM Medium |
Carefully add about 1/4 inch of Kovac's reagent to each of the 3 SIM agar tubes and observe.
1. Production of hydrogen sulfide (H2S)
2. Production of indole
SIM Medium
Record your results below (+ = positive; - = negative).
| Organism | Indole | Hydrogen Sulfide |
|---|---|---|
| Escherichia coli | ||
| Enterobacter cloacae | ||
| Proteus mirabilis |
E. Catalase Activity
| Video review - How to Interpret the Results of the Catalase Test |
Catalase is the name of an enzyme found in most bacteria which initiates the breakdown of hydrogen peroxide (H2O2) into water and free oxygen.
Record your results below (foaming = positive; no foaming = negative).
| Organism | Catalase Reaction |
|---|---|
| Staphylococcus aureus | |
| Streptococcus lactis |
PERFORMANCE OBJECTIVES FOR LAB 8
After the completion of this lab, the student will be able to complete the following objectives:
INTRODUCTION
1. State the chemical nature and function of enzymes.
2. Define endoenzyme and exoenzyme.
A. STARCH HYDROLYSIS
DISCUSSION
1. Describe a method of testing for starch hydrolysis and state how to interpret the results.
RESULTS
1. Interpret the results of starch hydrolysis on a Starch agar plate that has been inoculated, incubated, and flooded with iodine.
B. PROTEIN HYDROLYSIS
DISCUSSION
1. Describe a method of testing for casein hydrolysis and state how to interpret the results.
RESULTS
1. Interpret the results of casein hydrolysis on a Skim Milk agar plate after it has been inoculated and incubated.
C. FERMENTATION OF CARBOHYDRATES
DISCUSSION
1. Name the general end products which may be formed as a result of the bacterial fermentation of sugars and describe how these end products change the appearance of a broth tube containing a sugar, the pH indicator phenol red, and a Durham tube.
RESULTS
1. Interpret the carbohydrate fermentation results in tubes of Phenol Red Carbohydrate broth containing a Durham tube after it has been inoculated and incubated.
D. INDOLE AND HYDROGEN SULFIDE PRODUCTION
DISCUSSION
1. State the pathway for the breakdown of tryptophan to indole. 2. State the pathway for the detection of sulfur reduction in SIM medium.
3. State three reactions that may be tested for in SIM medium and describe how to interpret the results.
RESULTS
1. Interpret the hydrogen sulfide and indole results in a SIM medium tube after inoculation, incubation, and addition of Kovac's reagent.
E. CATALASE ACTIVITY
DISCUSSION
1. State the function of the enzyme catalase and describe a method of testing for catalase activity.
RESULTS
1. Interpret the results of a catalase test after adding hydrogen peroxide to a plate culture of bacteria.
SELF-QUIZ
Last updated: April, 2023
Microbiology Laboratory Manual by Gary E. Kaiser, PhD, Professor of Microbiology
is licensed under a Creative Commons Attribution 4.0 International License.
Please send comments and inquiries to Dr.
Gary Kaiser