Labs 12 and 13 deal with opportunistic and pathogenic glucose fermenting Gram-negative bacilli that are members of the bacterial family Enterobactereaceae, as well as glucose non-fermenting Gram-negative bacilli such as Pseudomonas and Acinetobacter. See Fig. 1.
Fig. 1: Flow Chart for Determining Whether or Not Your Unknown is a Glucose Fermenting or a Glucose Non-fermenting Gram-Negative Bacillus Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
A. ENTEROBACTERIACEAE: THE GLUCOSE-FERMENTING, GRAM-NEGATIVE, ENTERIC BACILLI
Bacteria belonging to the family Enterobacteriaceae are the most commonly encountered organisms isolated from clinical specimens. The Enterobacteriaceae is a large diverse family of bacteria belonging to the order Enterobacteriales in the class Gammaproteobacter of the phylum Proteobacter. Medically important members of this family are commonly referred to as glucose fermenting, Gram-negative, enteric bacilli, because they are Gram-negative bacilli that can ferment the sugar glucose. Many are normal flora of the intestinal tract of humans and animals while others infect the intestinal tract. Members of this family have the following characteristics in common:
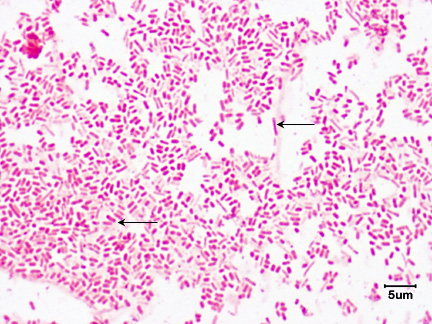
1. They are Gram-negative rods. See Fig. 2.
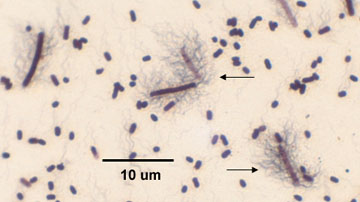
2. If motile, they possess a peritrichous arrangement of flagella. See Fig. 3.
3. They are facultative anaerobes.
4. With few exception, they are oxidase negative.
5. All species ferment the sugar glucose but otherwise vary widely in their biochemical characteristics.
6. Most reduce nitrates to nitrites.
Fig. 2: Escherichia coli, a Gram-negative Bacillus
Fig. 3: Flagella Stain of Proteus Showing Peritrichous Arrangement of Flagella
Note the bacterium is surrounded by flagella (arrow).Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
For further information on the Gram-negative cell wall, see the following CourseArc lecture lesson:
The Enterobacteriaceae represent a very large and diverse family of bacteria. Some of the more common clinically important genera of the family Enterobacteriaceae include:
| Salmonella | Citrobacter | Morganella |
| Shigella | Enterobacter | Yersinia |
| Proteus | Serratia | Edwardsiella |
| Escherichia | Klebsiella | Providencia |
Several genera of Enterobacteriaceae are associated with gastroenteritis and food-borne disease. These include:
- Salmonella.
- Shigella.
- Certain strains of Escherichia coli.
- Certain species of Yersinia.
All intestinal tract infections are transmitted by the fecal-oral route.
There are two species of Salmonella, Salmonella enterica and Salmonella bongori. Any infection caused by Salmonella is called a salmonellosis. Non-typhoidal Salmonella accounts for an estimated 520 cases per 100,000 population. CDC estimates that there are approximately 1,200,000 cases of non-typhoidal salmonellosis each year in the U.S resulting in 450 deaths. Since many different animals carry Salmonella in their intestinal tract, people usually become infected from ingesting improperly refrigerated, uncooked or undercooked poultry, eggs, meat, dairy products, vegetables, or fruit contaminated with animal feces.
Enteritis is the most common form of salmonellosis. Symptoms generally appear 6-48 hours after ingestion of the bacteria and include vomiting, nausea, non-bloody diarrhea, fever, abdominal cramps, myalgias, and headache. Symptoms generally last from 2 days to 1 week followed by spontaneous recovery. All species of Salmonella can cause bacteremia but S. enterica serotype Typhi, isolated only from humans, frequently disseminates into the blood causing a severe form of salmonellosis called typhoid fever. About 400 cases of typhoid fever occur each year in the U.S. but approximately 75% of these are acquired while traveling internationally.
Salmonella serotyping is a subtyping method of identification based on the identification of distinct cell wall, flagellar, and capsular antigens with known antiserum, as will be discussed in Lab 17. Salmonella serotypes Enteritidis and Typhimurium are the two most common serotypes in the United States, accounting for approximately 35 to 40% of all infections confirmed by laboratory culture. As mentioned above, S. enterica serotype Typhi is responsible for typhoid fever.
Any Shigella infection is called a shigellosis. Unlike Salmonella, which can infect many different animals, Shigella only infects humans and other higher primates. There are approximately 14,000 laboratory cases of shigellosis a year reported in the US with an estimated 450,000 total cases and 70 deaths.
Shigellosis frequently starts with a watery diarrhea, fever, and abdominal cramps but may progress to dysentery with scant stools containing blood, pus, and mucus. The incubation period is 1-3 days. Initial profuse watery diarrhea typically appears first as a result of enterotoxin. Within 1-2 days this progresses to abdominal cramps, with or without bloody stool. Classic shigellosis presents itself as lower abdominal cramps and stool abundant with blood and pus develops as the Shigella invade the mucosa of the colon.
Escherichia coli is one of the dominant normal flora in the intestinal tract of humans and animals. Some strains, however, can cause infections of the intestines while others are capable of causing infections outside the intestines. Extraintestinal pathogenic E. coli cause such opportunistic infections as urinary tract infections, wound infections, and septicemia and will be discussed in greater detail below. Intestinal or diarrheagenic E. coli cause infections of the intestinal tract. Diarrheagenic E. coli include:
- Enterotoxigenc E. coli (ETEC) produce enterotoxins that cause the loss of sodium ions and water from the small intestines resulting in a watery diarrhea. It is an important cause of diarrhea in impoverished countries. ETEC accounts for between 11-15% of cases of traveler's diarrhea in persons visiting developing countries and 30-45% of cases among those visiting Mexico. There are approximately 80,000 cases of ETEC a year in the U.S.
- Enteropathogenic E. coli (EPEC) causes an endemic diarrhea in in impoverished countries, especially in infants younger than 6 months of age. The bacterium disrupts the normal microvilli on the epithelial cells of the small intestines resulting in maladsorbtion and diarrhea. They do not produce enterotoxin or shiga toxin and are not invasive. It is rare in industrialized countries.
- Enteroaggregative E. coli (EAEC) is a cause of endemic diarrhea in children in impoverished countries and industrialized countries. EAEC causes around 30% of cases of traveler's diarrhea. It is also responsible for a persistent diarrhea in people infected with HIV. It probably causes diarrhea by adhering to mucosal epithelial cells of the small intestines and interfering with their function.
- Enteroinvasive E. coli (EIEC) invade and kill epithelial cells of the colon usually causing a watery diarrhea but sometimes progressing to a dysentery-type syndrome with blood in the stool. It occurs mostly in impoverished countries and is rare in industrialized countries.
- Shiga toxin-producing E. coli (STEC) such as E. coli 0157:H7, produce a shiga toxin that kills epithelial cells of the colon causing hemorrhagic colitis, a bloody diarrhea. In rare cases, the shiga toxin enters the blood and is carried to the kidneys where, usually in children, it damages vascular cells and causes hemolytic uremic syndrome. E. coli 0157:H7 is thought to cause more than 20,000 infections and up to 250 deaths per year in the U.S.
Several species of Yersinia, such as Y. enterocolitica and Y. pseudotuberculosis are also causes of diarrheal disease.
Many other genera of the family Enterobacteriaceae are normal microbiota of the intestinal tract and are considered opportunistic pathogens. The most common genera of Enterobacteriaceae causing opportunistic infections in humans include the following:
- Escherichia coli.
- Proteus.
- Enterobacter.
- Klebsiella.
- Citrobacter.
- Serratia.
They act as opportunistic pathogens when they are introduced into body locations where they are not normally found, especially if the host is debilitated or immunosuppressed. They all cause the same types of opportunistic infections, namely:
- Urinary tract infections.
- Wound infections.
- Pneumonia.
- Septicemia.
These normal flora Gram-negative bacilli, along with Gram-positive bacteria such as Enterococcus species (see Lab 14) and Staphylococcus species (see Lab 15), are among the most common causes of health-care-associated infections (formerly called nosocomial infections).
According to the Centers for Disease Control and Prevention (CDC) Health-care-associated infection's website, "In American hospitals alone, health-care-associated infections account for an estimated 1.7 million infections and 99,000 associated deaths each year. Of these infections:
- 32 percent of all health-care-associated infection are urinary tract infections (UTIs.)
- 22 percent are surgical site infections.
- 15 percent are pneumonia (lung infections.)
- 14 percent are bloodstream infections.
Most patients who have healthcare-associated infections are predisposed to infection because of invasive supportive measures such as urinary catheters, intravascular lines, and endotracheal intubation.
By far, the most common Gram-negative bacterium causing nosocomial infections is Escherichia coli. E. coli causes between 70 and 90% of both upper and lower urinary tract infections (UTIs). It is also a frequent cause of abdominal wound infections and septicemia. Depending on the facility, E. coli is responsible for between 12% and 50% of all healthcare-associated infections.
However, according to a 2008 study, Enterobacteriaceae other than E. coli were responsible for 7 of the 10 most common Gram-negative organisms isolated from urinary tract, respiratory tract, and bloodstream infections from intensive care unit patients between 2002 and 2008 in the United States. These include Klebsiella pneumoniae (15%), Enterobacter cloacae (9%), Serratia marcescens (6%), Enterobacter aerogenes (4%), Proteus mirabilis (4%), Klebsiella oxytoca (3%), and Citrobacter freundii (2%). Furthermore, the National Healthcare Safety Network reported K. pneumoniae (6%) , Enterobacter spp. (5%), and K. oxytoca (2%) among the top 10 most frequently isolated health care-associated infections between the years between 2006 and 2007.
1. Urinary Tract Infections
The most common infection caused by opportunistic Enterobacteriaceae is a urinary tract infection (UTI). UTIs account for more than 8, 000,000 physician office visits per year in the U.S and as many as 100,000 hospitalizations. Among the non-hospitalized and non-debilitated population, UTIs are more common in females because of their shorter urethra and the closer proximity between their anus and the urethral opening. (Over 20 percent of women have recurrent UTIs.) However, anyone can become susceptible to urinary infections in the presence of predisposing factors that cause functional and structural abnormalities of the urinary tract. These abnormalities increase the volume of residual urine and interfere with the normal clearance of bacteria by urination. Such factors include prostate enlargement, sagging uterus, expansion of the uterus during pregnancy, paraplegia, spina bifida, scar tissue formation, and catheterization. Between 35 and 40 percent of all nosocomial infections, about 900,000 per year in the U.S., are UTIs and are usually associated with catheterization.
E. coli and Staphylococcus saprophyticus (a Gram-positive staphylococcus that will be discussed in Lab 15) cause around 90 percent of all uncomplicated UTIs. Most of the remaining uncomplicated UTIs are caused by other Gram-negative enterics such as Proteus mirabilis and Klebsiella pneumoniae or by Enterococcus faecalis (a Gram-positive streptococcus that will be discussed in Lab 14). E. coli is responsible for more than 50 percent of healthcare-associated UTIs. Other causes of hospital-acquired UTIs include other species of Enterobacteriaceae (such as Proteus, Enterobacter, and Klebsiella), Pseudomonas aeruginosa (discussed below), Enterococcus species (discussed in lab 14), Staphylococcus saprophyticus (discussed in Lab 15), and the yeast Candida (discussed in lab 9).
The traditional laboratory culture standard for a UTI has been the presence of more than 100,000 CFUs (colony-forming units; see Lab 4) per milliliter (ml) of midstream urine, or any CFUs from a catheter-obtained urine sample. More recently, this has been modified and counts of as few as 1000 colonies of a single type per ml or as little as 100 coliforms per ml are now considered as indicating a UTI.
2. Wound Infections
Wound infections are due to fecal contamination of external wounds or a result of wounds that cause trauma to the intestinal tract, such as surgical wounds, gunshot wounds, and knife wounds. In the latter case, fecal bacteria get out of the intestinal tract and into the peritoneal cavity causing peritonitis and formation of abscesses on the organs found in the peritoneal cavity.
3. Pneumonia
Although they sometimes cause pneumonia, the Enterobacteriaceae account for less than 5% of the bacterial pneumonias requiring hospitalization.
4. Bloodstream Infections
Gram-negative septicemia is a result of these opportunistic Gram-negative bacteria getting into the blood. They are usually introduced into the blood from some other infection site, such as an infected kidney, wound, or lung. Looking at patients that develop septic shock:
- Lower respiratory tract infections are the source in about 25% of patients.
- Urinary tract infections are the source in about 25% of patients.
- Soft tissue infections are the source in about 15% of patients.
- Gastrointestinal infections are the source in about 15% of patients.
- Reproductive tract infections are the source in about 10% of patients.
- Foreign bodies (intravascular lines, implanted surgical devices, etc.) are the source in about 5% of patients.
There are approximately 750,000 cases of septicemia per year in the U.S. and 200,000 cases of septic shock. Septic shock results in approximately 100,000 deaths per year in the U.S. Approximately 45 percent of the cases of septicemia are due to Gram-negative bacteria. Klebsiella, Proteus, Enterobacter, Serratia, and E. coli, are all common Enterobacteriaceae causing septicemia. (Another 45 percent are a result of Gram-positive bacteria (see Labs 14 and 15) and 10 percent are due to fungi, mainly the yeast Candida (see Lab 9).
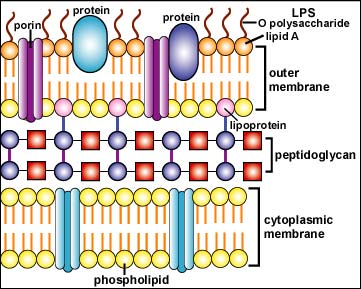
In the outer membrane of the Gram-negative cell wall, the lipid A moiety of the lipopolysaccharide (LPS) functions as an endotoxin (see Fig 4). Endotoxin indirectly harms the body when massive amounts are released during severe Gram-negative infections. This, in turn, causes an excessive cytokine response.
Fig. 4: Structure of a Gram-Negative Cell Wall
The Gram-negative cell wall is composed of a thin, inner layer of peptidoglycan and an outer membrane consisting of molecules of phospholipids, lipopolysaccharides (LPS), lipoproteins and sutface proteins.The lipopolysaccharide consists of lipid A and O polysaccharide.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
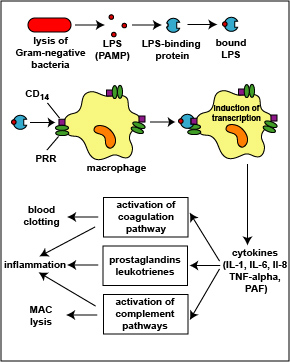
1. The LPS released from the outer membrane of the Gram-negative cell wall first binds to a LPS-binding protein circulating in the blood and this complex, in turn, binds to a receptor molecule (CD14) found on the surface of body defense cells called macrophages (see Fig. 5) located in most tissues and organs of the body.
2. This is thought to promote the ability of the toll-like receptor TLR-4 to respond to the LPS, triggering the macrophages to release various defense regulatory chemicals called cytokines, including tumor necrosis factor-alpha (TNF-alpha), interleukin-1 (IL-1), interleukin-6 (IL-6), and interleukin-8 (IL-8), and platelet-activating factor (PAF). The cytokines then bind to cytokine receptors on target cells and initiate inflammation and activate both the complement pathways and the coagulation pathway (see Fig. 5).
Fig. 5: Physiologic Action of Lipopolysaccharide (LPS) from the Gram-Negative Cell Wall
The lysis of Gram-negative bacteria causes them to release lipopolysaccharide (LPS; endotoxin) from the outer membrane of their cell wall. The LPS binds to a LPS-binding protein circulating in the blood and this complex, in turn, binds to a receptor molecule (CD14) found on the surface of body defense cells called macrophages. This is thought to promote the ability of the toll-like receptor TLR-4 to respond to the LPS, triggering the macrophage to release various defense regulatory chemicals called cytokines, including IL-1, IL-6, IL-8, TNF-alpha, and PAF. The cytokines then bind to cytokine receptors on target cells and initiate inflammation and activate both the complement pathways and the coagulation pathway.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
3. The complex of LPS and LPS binding protein can also attach to molecules called CD14 on the surfaces of phagocytic white blood cells called neutrophils causing them to release proteases and toxic oxygen radicals for extracellular killing. Chemokines such as interleukin-8 (IL-8) also stimulate extracellular killing. In addition, LPS and cytokines stimulate the synthesis of a vasodilator called nitric oxide.
During minor local infections with few bacteria present, low levels of LPS are released leading to moderate cytokine production by the monocytes and macrophages and in general, promoting body defense by stimulating inflammation and moderate fever, breaking down energy reserves to supply energy for defense, activating the complement pathway and the coagulation pathway, and generally stimulating immune responses (see Fig. 5 above). Also as a result of these cytokines, circulating phagocytic white blood cells such as neutrophils and monocytes stick to the walls of capillaries, squeeze out and enter the tissue, a process termed diapedesis. The phagocytic white blood cells such as neutrophils then kill the invading microbes with their proteases and toxic oxygen radicals.
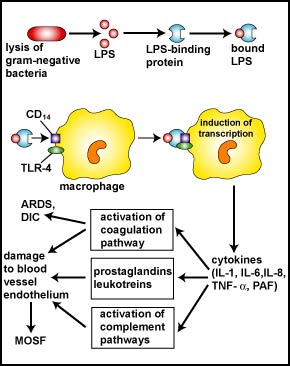
However, during severe systemic infections with large numbers of bacteria present, high levels of LPS are released resulting in excessive cytokine production by the monocytes and macrophages and this can harm the body (see Fig. 6). In addition, neutrophils start releasing their proteases and toxic oxygen radicals that kill not only the bacteria, but the surrounding tissue as well. Harmful effects include high fever, hypotension, tissue destruction, wasting, acute respiratory distress syndrome (ARDS), disseminated intravascular coagulation (DIC), and damage to the vascular endothelium resulting in shock, multiple system organ failure (MOSF), and often death.
Fig. 6: Harmful Effects of Lipopolysaccharide (LPS; Endotoxin) Released from the Gram-Negative Cell Wall
The lysis of Gram-negative bacteria causes them to release lipopolysaccharide (LPS; endotoxin) from the outer membrane of their cell wall. The LPS binds to a LPS-binding protein circulating in the blood and this complex, in turn, binds to a receptor molecule (CD14) found on the surface of body defense cells called macrophages. This triggers the macrophages to release various defense regulatory chemicals called cytokines, including IL-1, IL-6, IL-8, TNF-alpha, and PAF. The cytokines then bind to cytokine receptors on target cells stimulating the production of inflammatory mediators such as prostaglandins and leukotrienes as well as activating both the complement pathways and the coagulation pathway. Excessive production of clotting factors may lead to ARDS and DIC while an overproduction of prostaglandins, leukotrienes, and complement proteins can damage the vascular endothelium resulting in shock and MOSF. (LPS, lipopolysaccharide; IL-1, interleukin-1; IL-6, interleukin-6; IL-8, interleukin-8, TNF-alpha, tumor necrosis factor-alpha; PAF, platelet-activating factor; ARDS, acute respiratory distress syndrome; DIC, disseminated intravascular coagulation; MOSF, multiple organ system failure.)Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
This excessive inflammatory response is referred to as Systemic Inflammatory Response Syndrome or SIRS. Death is a result of what is called the shock cascade. The sequence of events is as follows:
The release of excessive levels of inflammatory cytokines in response to PAMPs binding to PRRs during a systemic infection results in:
1. A drop in blood volume or hypovolemia. This is caused by the following events:
a. Extracellular killing by neutrophils damages the capillary walls resulting in blood and plasma leaving the bloodstream and entering the surrounding tissue.
b. Depletion of clotting factors during disseminated intravascular coagulation (DIC) can lead to hemorrhaging as the capillaries are damaged.
c. Prolonged vasodilation results in plasma leaving the bloodstream and entering the surrounding tissue.
2. A drop in blood pressure or hypotension. This is a result of the following events:
a. Prolonged vasodilation causes decreased vascular resistance within blood vessels decreases blood pressure.
b. High levels of TNF, inhibit vascular smooth muscle tone and myocardial contractility decreasing the ability of the heart to pump blood throughout the body.
c. Hypovolemia from capillary damage, plasma leakage, and hemorrhaging.
3. The inability to deliver nutrients and oxygen to body cells or hypoperfusion. This is a result of the following events:
a. Activation of the blood coagulation pathway can cause clots called microthrombi to form within the blood vessels throughout the body causing disseminated intravascular coagulation (DIC) which blocks the flow of blood through the capillaries and, as mentioned above, depletion of clotting factors can lead to hemorrhaging in many parts of the body.
b. Increased capillary permeability as a result of vasodilation in the lungs, as well as neutrophil-induced injury to capillaries in the alveoli leads to acute inflammation, pulmonary edema, and loss of gas exchange in the lungs (acute respiratory distress syndrome or ARDS). As a result, the blood does not become oxygenated.
c. Hypovolemia decreases the volume of circulating blood and leads to hypotension.
d. Hypotension decreases the pressure needed to deliver blood throughout the body.
6. Hypoperfusion in the liver can result in a drop in blood glucose level from liver dysfunction. Glucose is needed for ATP production during glycolysis and aerobic respiration. A drop in glucose levels can result in decreased ATP production and insufficient energy for cellular metabolism.
7. The lack of oxygen delivery as a result of hypoperfusion causes cells to switch to fermentation for energy production. The lactic acid end products of fermentation may lead to lactic acidosis and a blood pH to low for the functioning of the enzymes involved in cellular metabolism. This can result in irreversible cell death.
Collectively, this can result in :
- end-organ ischemia: a restriction in blood supply that results in damage or dysfunction of tissues or organs,
- multiple system organ failure (MSOF),
- death.
Both pili and surface proteins in the Gram-negative cell wall function as adhesins, allowing the bacterium to adhere intimately to host cells and other surfaces in order to colonize and resist flushing. Some Gram-negative bacteria also produce invasins, allowing some bacteria to invade host cells. Motility, capsules, biofilm formation, and exotoxins also play a role in the virulence of some Enterobacteriaceae.
For further information on virulence factors associated with various Enterobacteriaceae, see my CourseArc Lecture Lessons listed below:
- Gram-Negative PAMPs
- The Ability to Adhere to Host Cells and Resist Physical Removal
- The Ability to Invade Host Cells
- The Ability to Contact Host Cells
- The Ability to Resist Phagocytic Engulfment (Attachment and Ingestion) and Antibacterial Peptides
- The Ability to Resist Phagocytic Destruction
- Type III toxins (A-B Toxins and Other Toxins that Interfere with Host Cell Function)
- Type II toxins (Toxins that Damage Host Cell Membranes)
Many of the Enterobacteriaceae also possess R (resistance) plasmids. These plasmids are small pieces of circular non-chromosomal DNA that may code for multiple antibiotic resistance In addition, the plasmid may code for a sex pilus, enabling the bacterium to pass R plasmids to other bacteria by conjugation. Between 50 and 60 percent of the bacteria causing healthcare-associated infections are antibiotic resistant.
For further information on bacterial resistance to antibiotics, see the following CourseArc lessons:
The identification of lactose-fermenting Gram-negative rods belonging to the bacterial family Enterobacteriaceae (bacteria commonly referred to as coliforms) in water is often used to determine if water has been fecally contaminated and, therefore, may contain disease-causing pathogens transmitted by the fecal-oral route. The procedure for this is given in Appendix E.
B. PSEUDOMONAS AND OTHER GLUCOSE NON-FERMENTING GRAM-NEGATIVE BACILLI
Glucose non-fermenting Gram-negative bacilli refer to Gram-negative bacilli or coccobacilli that cannot ferment the sugar glucose. The glucose non-fermenting Gram-negative bacilli are often normal inhabitants of soil and water. They may cause human infections when they colonize immunosuppressed individuals or gain access to the body through trauma. However, less than one-fifth of the Gram-negative bacilli isolated from clinical specimens are glucose non-fermenting bacilli. By far, the most common Gram-negative, glucose non-fermenting rod that causes human infections is Pseudomonas aeruginosa. Pseudomonas belongs to the family Pseudomonadaceae in the order Pseudomonadales in the class Gammaproteobacter of the phylum Proteobacter.
Pseudomonas aeruginosa is also an opportunistic pathogen. It is a common cause of nosocomial infections and can be found growing in a large variety of environmental locations. In the hospital environment, for example, it has been isolated from drains, sinks, faucets, water from cut flowers, cleaning solutions, medicines, and even disinfectant soap solutions. It is especially dangerous to the debilitated or immunocompromised patient.
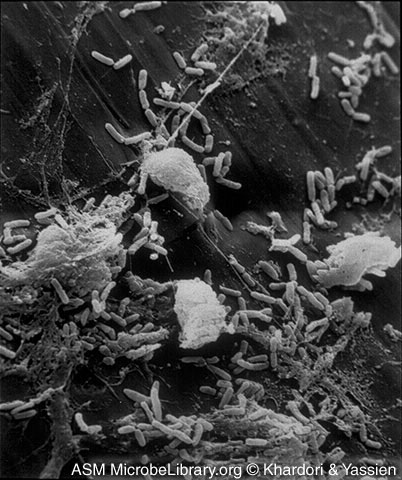
Like the opportunistic Enterobacteriaceae, Pseudomonas is a Gram-negative rod, it is frequently found in small amounts in the feces, and it causes similar opportunistic infections: urinary tract infections, wound infections, pneumonia, and septicemia. P aeruginosa is the fourth most commonly isolated nosocomial pathogen, accounting for 10% of all hospital-acquired infections. P. aeruginosa is responsible for 12 percent of hospital-acquired urinary tract infections, 16 percent of nosocomial pneumonia cases, and 10 percent of the cases of septicemia. In addition, P. aeruginosa is a significant cause of burn infections with a 60 percent mortality rate. It also colonizes and chronically infects the lungs of people with cystic fibrosis. Like other opportunistic Gram-negative bacilli, Pseudomonas aeruginosa also releases endotoxin and frequently possesses R-plasmids. A number of other species of Pseudomonas have also been found to cause human infections. See Fig. 7 to view an electron micrograph of Pseudomonas aeruginosa colonizing a vascular catheter.
Fig. 7: Scanning Electron Micrograph of Pseudomonas aeruginosa Colonizing a Vascular Catheter
The microcolonies of Pseudomonas are attaced to the catheter by a glycocalyx.Microcolonies of Bacteria Growing on a Catheter Segment, Part I. Mahmoud Yassien and Nancy Khardori, authors.
Licensed for use, ASM MicrobeLibrary.
Other glucose non-fermenting Gram-negative bacilli that are sometimes opportunistic pathogens in humans include Acinetobacter, Aeromonas, Alcaligenes, Eikenella, Flavobacterium, and Moraxella.
Acinetobacter has become a frequent cause of nosocomial wound infections, pneumonia, and septicemia. The bacterium has become well known as a cause of infections among veterans of the wars in Iraq and Afghanistan and is becoming a growing cause of nosocomial infections in the U.S. Acinetobacter is thought to have been contracted in field hospitals in Iraq and Afghanistan and subsequently carried to veteran's hospitals in the U.S. Because most species are multiple antibiotic resistant, it is often difficult to treat. Acinetobacter is commonly found in soil and water, as well as on the skin of healthy people, especially healthcare personnel. Although there are numerous species of Acinetobacter that can cause human disease, Acinetobacter baumannii accounts for about 80% of reported infections.
| Medscape articles on infections associated with organisms mentioned in this lab exercise. Registration to access this website is free.
|
Students will be assigned either Case Study 1A, 1B, or 1C to do today. All students will do Case Study 2 as part of the results next time.
Case Study #1A
A 26-year-old female presents to her doctor complaining of 2 days of increased urinary frequency, dysuria, and sensation of incomplete voiding. Her abdominal exam indicates mild suprapubic tenderness. Her blood pressure is normal and she does not have fever, chills, costovertebral angle (CVA) tenderness, or vaginal discharge. She reports that she became sexually active with her new boyfriend one month ago. She and her boyfriend have sexual intercourse 3-4 times a week. She is using a combination of a diaphragm and spermicide for contraception. She is otherwise healthy. A microscopic examination of her centrifuged urine shows 9 white blood cells and 15 bacteria per high-power microscopic field. A urine dipstick shows a positive leukocyte esterase test and a positive nitrite test.
Assume that your unknown is from the urine of this patient.
Case Study #1B
A 90-year-old woman resides at an area nursing home. She shows signs of mild dementia, and because of severe arthritis and requiring a walker for ambulation, sits in a chair most of the day. She has not used any form of estrogen in at least 30 years. She also has a history of 4-5 confirmed urinary tract infections per year. This morning, her caregiver is unable to coax the patient out of her bed. She seems confused and disoriented. Vital signs reveal tachycardia in the 120’s, respirations at 24/min, and a blood pressure of 78/49. She is immediately taken to an ER for evaluation. A CT of the abdomen and a chest x-ray appear normal. She has a WBC count of 2300/μL. She continues to exhibits marked confusion compared to her baseline and is exhibiting anxiety. Urine and blood samples are taken and sent for culture and sensitivity
.
Assume your unknown is from both a urine sample and a blood sample.
Case Study #1C
A 79-year-old man living in a nursing home has COPD, a lifetime history of heavy smoking, and hypertension. His caregivers note that he is exhibiting rigor, has a temperature of 103°F, and lacks his normal alertness. Vital signs include a blood pressure is 165/90, a pulse of 128 beats per minute, a respiratory rate of 32 breaths per minute, and a pulse oximetry on room air of 80%. He is transferred to an acute care facility where a chest X-ray reveals a right lower lobe infiltrate and his white blood cell count is 18,000/μL with a marked left shift. He has thick, foul-smelling yellow-green sputum.
Assume you unknown is from the sputum sample.
CAUTION: TREAT EACH UNKNOWN AS A PATHOGEN!. Inform your instructor of any spills or accidents. WASH AND SANITIZE YOUR HANDS WELL before leaving the lab.
MATERIALS
Gibson Oxidase Test Swab and either a plate of MacConkey agar or a plate of Cetrimide agar, and an EnterPluri-Test
| How to Chemically Fix a Microscope Slide with Methanol |
| How to Prepare a Slide for Staining when using Bacteria from an Agar Culture |
| How to Make a Gram Stain |
| A Review of the Critical Decolorization Step of the Gram Stain |
Keep in mind that organisms other than the Enterobacteriaceae and Pseudomonas can cause these infections, so in a real clinical situation other lab tests and cultures for bacteria other than those upon which this lab is based would also be done.
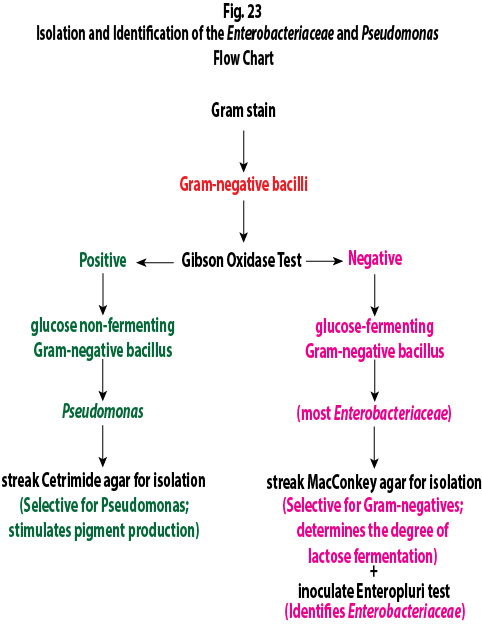
Flow chart of the tests you will be using today. See Fig. 1
Fig. 1: Flow Chart for Determining Whether or Not Your Unknown is a Glucose Fermenting or a Glucose Non-fermenting Gram-Negative Bacillus Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
1. Perform a Gram stain on your unknown. Remember that the concentration of bacteria on slides prepared from taking bacteria off a petri plate tend to be much greater than those prepared by taking bacteria out of a broth culture, so be careful not to under decolorize. Continue decolorizing until the purple just stops flowing off of the bacterial smear, then wash with water.
Record the results of your Gram stain in the Gram stain section of Lab 13.
2. If you have a Gram-negative bacillus, determine if it is a glucose fermenting Gram-negative bacillus like most Enterobacteriaceae or a glucose non-fermenting Gram-negative bacillus such as Pseudomonas by performing an oxidase test as follows:
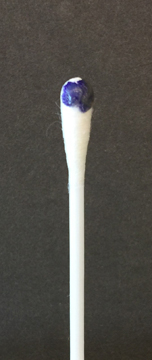
To perform the oxidase test, touch the oxidase test swab to a colony on your unknown plate. An oxidase-positive reaction will appear purple to black within 10 seconds as shown in Fig. 8A. An oxidase-negative reaction will result in no color change as shown in Fig. 8B. Ignore any color development occurring after 60 seconds.
Record your oxidase test results in the Oxidase test section of Lab 13.
Fig. 8A: A Positive Oxidase Test
Fig. 8B: A Negative Oxidase Test
Touch the Gibson Oxidase Test swab to a colony. An oxidase-positive reactions will appear purple to black within 10 seconds as shown in the above image. Disregard any color development that appears after 60 seconds.
Touch the Gibson Oxidase Test swab to a colony. An oxidase-positive reactions will appear purple to black within 10 seconds as shown in the above image. No change in color is negative. Disregard any color development that appears after 60 seconds.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
3. If your unknown is oxidase-negative, usually indicating a glucose fermenting Gram-negative bacillus, do the following inoculations:
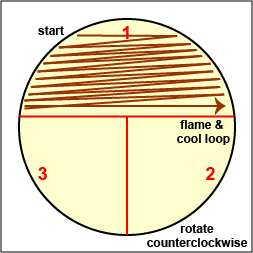
a. Streak your unknown for isolation on a plate of MacConkey agar, a selective medium used for the isolation of non-fastidious Gram-negative rods and particularly members of the family Enterobacteriaceae, as illustrated in Fig. 9 through 9C. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section.
Fig. 9A: Streaking For Isolation, Step-1
Fig. 9B: Streaking For Isolation, Step-2
Fig. 9C: Streaking For Isolation, Step-3
Streak the inoculum over area "1" as shown above. Then flame the loop and cool it by sticking it into the edge of the agar. Rotate the plate 90 degrees counterclockwise. Rotate the plate so area "1" is on your left. Drag your stetile inoculating loop through area "1" two times and spread out over area "2" as shown above. Then flame the loop and cool it by sticking it into the edge of the agar. Rotate the plate 90 degrees counterclockwise. 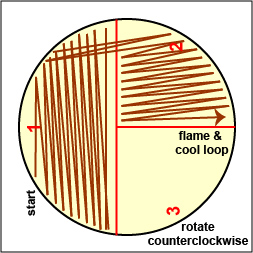
Rotate the plate so that area "2" is on your left. Drag your sterile inoculating loop through area "2" two times and spread the inoculum on area "2" over area "3." Sterilize the loop. Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
b. Inoculate an EnterPluri-Test as follows:
1. Remove both caps of the EnteroPluri-Test and with the straight end of the inoculating wire, pick off the equivalent of a colony from your unknown plate. A visible inoculum should be seen on the tip and side of the wire.
2. Inoculate the EnteroPluri-Test by grasping the bent-end of the inoculating wire, twisting it, and withdrawing the wire through all 12 compartments using a turning motion.
3. Reinsert the wire into the tube (use a turning motion) through all 12 compartments until the notch on the wire is aligned with the opening of the tube. (The tip of the wire should be seen in the citrate compartment.) Break the wire at the notch by bending. Do not discard the wire yet.
4. Using the broken off part of the wire, punch holes through the cellophane which covers the air inlets located on the rounded side of the last 8 compartments. Your instructor will show you their correct location. Discard the broken off wire in the disinfectant container.
5. Replace both caps and incubate the EnteroPluri-Test on its flat surface at 36°- 37°C for 18-24 hours.
4. If your unknown is oxidase-positive, usually indicating a glucose non-fermenting Gram-negative bacillus, do the following inoculation:
a. Streak your unknown for isolation on a plate of Cetrimide agar, a selective and differential medium for Pseudomonas, as illustrated in Fig. 9A through 9C above. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section.
Note that MacConkey agar can also be used to isolate Pseudomonas but we are using the Cetrimide agar today because it enables us to detect the production of the blue to green water-soluble pigment by Pseudomonas aeruginosa, as well as the production of fluorescein.
You will also inoculate an EnteroPluri-Test for practice only, but keep in mind that the EnteroPluri-Test is used to identify Enterobacteriaceae, not Pseudomonas.
After receiving a baby chicken for Easter, a 7-year-old boy is taken to the emergency room with symptoms of vomiting, nausea, non-bloody diarrhea, abdominal cramps, and a temperature of 100°F. A complete blood count (CBC) shows the WBC count to be within the reference range.
This XLD agar plate and this EnteroPluri-Test are from a stool culture from this patient.
CAUTION: TREAT THE UNKNOWN AS A PATHOGEN!. Inform your instructor of any spills or accidents. WASH AND SANITIZE YOUR HANDS WELL before leaving the lab.
MATERIALS
Demonstration XLD agar plate and EnteroPluri-Test
PROCEDURE (to be done in groups of 3)
1. Observe the following demonstrations shown below and identify the causative bacterium:
a. Fig. 10: An XLD agar plate. XLD agar is a selective medium used for isolating and differentiating Gram-negative enteric bacteria, especially intestinal pathogens such as Salmonella and Shigella.
b. Fig. 11: An EnteroPluri-Test.
Fig. 10: Your Unknown Growing on XLD Agar for Case Study #2
Fig. 11: Your Unknown Growing in an EnteroPluri Test for Case Study #2

Photo courtesy of Nathan Reading, Halesowen, UK Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
2. Record your results in the Results section of Lab 13.
C. Lab Tests Used as Part of Today's Lab
To isolate Enterobacteriaceae and Pseudomonas, specimens from the infected site are plated out on any one of a large number of selective and differential media such as EMB agar, Endo agar, Deoxycholate agar, MacConkey agar, Hektoen Enteric agar, and XLD agar. We will look at three of these.
1. MacConkey Agar
Videos review: How to Interpret the Results of MacConkey Agar
MacConkey agar is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives. See Fig. 12).
Fig. 12: An Uninoculated Plate of MacConkey Agar
MacConkey agar is a selective medium used for the isolation of non-fastidious Gram-negative rods, particularly members of the family Enterobacteriaceae and the genus Pseudomonas, and the differentiation of lactose fermenting from lactose non-fermenting Gram-negative bacilli. MacConkey agar contains the dye crystal violet well as bile salts that inhibit the growth of most Gram-positive bacteria but do not affect the growth of most Gram-negatives. Photograph from From MicrobeLibrary.org
Courtesy of David Miller and Patrick Hanley, Hartwick College, Oneonta, NY.
If the Gram-negative bacterium ferments the sugar lactose in the medium, the acid end products lower the pH of the medium. The neutral red in the agar turns red in color once the pH drops below 6.8. As the pH drops, the neutral red is absorbed by the bacteria, causing the colonies to appear bright pink to red.
- Strong fermentation of lactose with high levels of acid production by the bacteria causes the colonies and confluent growth to appear bright pink to red. The resulting acid, at high enough concentrations, can also causes the bile salts in the medium to precipitate out of solution causing a pink precipitate (cloudiness) to appear in the agar surrounding the growth. See Fig. 13).
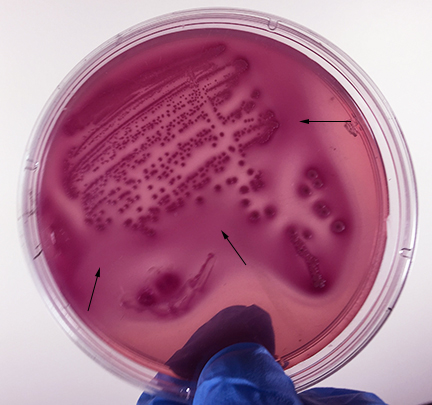
Fig. 13: Escherichia coli Growing on MacConkey Agar
Strong fementation of lactose with high levels of acid production by the bacteria causes the colonies and confluent growth to appear bright pink to red. The resulting acid, at high enough concentrations, can also causes the bile salts in the medium to precipitate out of solution causing a pink precipitate (cloudiness to appear around the the growth (arrows). Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
- Weak fermentation of lactose by the bacteria causes the colonies and confluent growth to appear pink to red, but without the precipitation of bile salts there is no pink halo around the growth. See Fig. 14.
Fig. 15: Klebsiella (Enterobacter) aerogenes Growing on MacConkey Agar
Weak fermentation of lactose by the bacteria causes the colonies and confluent growth to appear pink to red, but without the precipitation of bile salts there is no pink halo around the growth (arrow). Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
- If the bacteria do not ferment lactose, the colonies and confluent growth appear colorless and the agar surrounding the bacteria remains relatively transparent. See Fig. 16A and Fig. 16B.
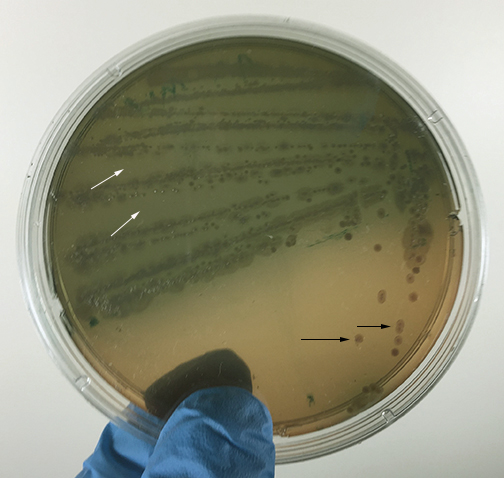
Fig. 16A: Proteus mirabilis Growing on MacConkey Agar
Fig. 16B: Pseudomonas aeruginosa Growing on MacConkey Agar
If the bacteria do not ferment lactose, the colonies and confluent growth appear colorless and the agar surrounding the bacteria remains relatively transparent (arrow). If the bacteria do not ferment lactose, the colonies and confluent growth appear relatively colorless and the agar surrounding the bacteria remains relatively transparent (black arrows). The green, water-soluble pigment produced by Pseudomonas aeruginosa (white arrows) is also evident here. Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Typical results for our strains of Enterobacteriaceae and Pseudomonas aeruginosa on MacConkey agar is as follows:
A. Strong fermentation of lactose
Escherichia coli
B. Weak fermentation of lactose
Klebsiella pneumoniae, Klebsiella aerogenes (formerly known as Enterobacter aerogenes), Enterobacter cloacae.
C. No fermentation of lactose
Proteus mirabilis, Proteus vulgaris, Serratia marcescens, Salmonella enterica, Pseudomonas aeruginosa.
2. XLD Agar
Xylose Lysine Desoxycholate (XLD) agar is used for isolating and differentiating Gram-negative enteric bacteria, especially intestinal pathogens such as Salmonella and Shigella. XLD agar contains sodium desoxycholate, which inhibits the growth of Gram-positive bacteria but permits the growth of Gram-negatives. It also contains the sugars lactose and sucrose, the amino acid L-lysine, sodium thiosulfate, and the pH indicator phenol red. Results can be interpreted as follows:
- If the Gram-negative bacterium ferments lactose and/or sucrose, acid end products will be produced and cause the colonies and the phenol red in the agar around the colonies to turn yellow (see Fig. 17).
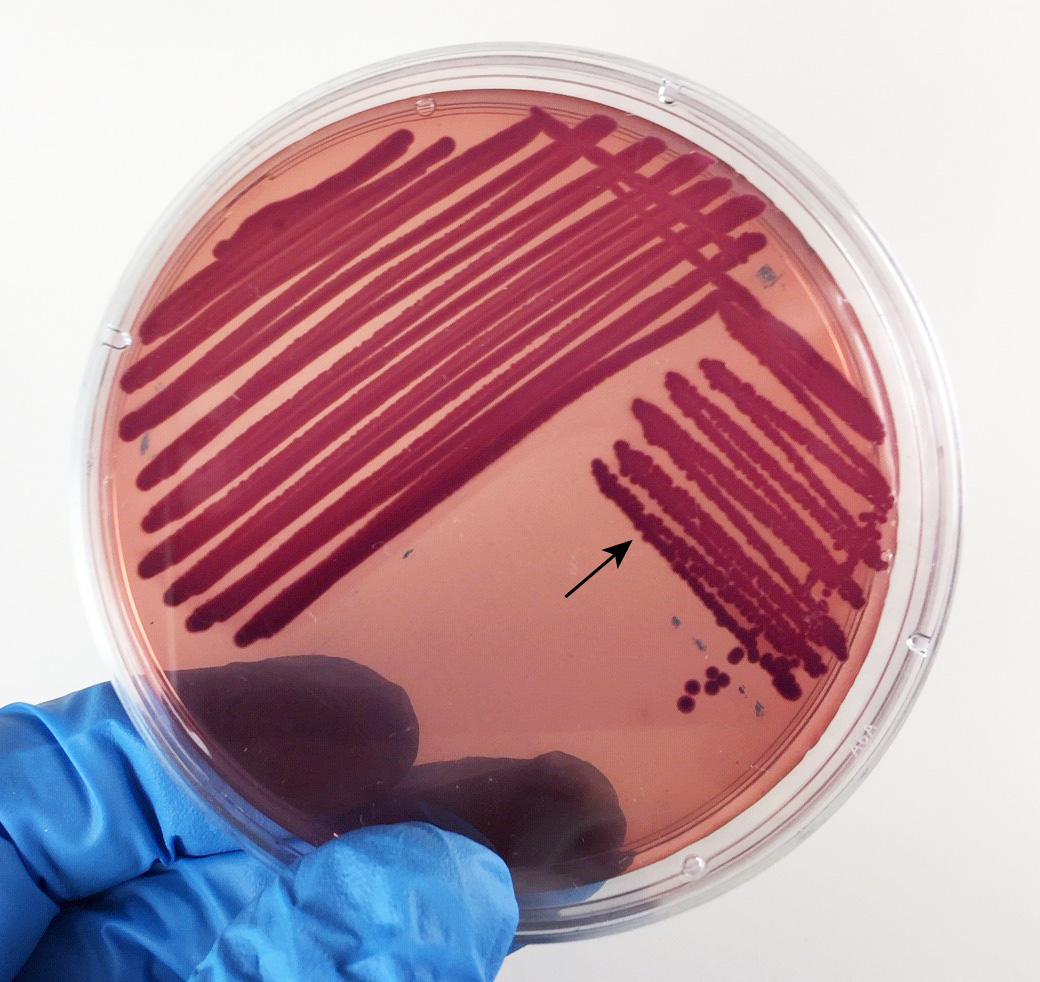
Fig. 17: Escherichia coli Growing on XLD Agar
Acid from fermentation lowers the pH and turns the phenol red from red (alkaline) to yellow (acid). No hydrogen sulfide is produced. Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
- If lactose and sucrose are not fermented by the bacterium but the amino acid lysine is decarboxylated, ammonia, an alkaline end product will cause the phenol red in the agar around the colonies to turn a deeper red (see Fig. 18).
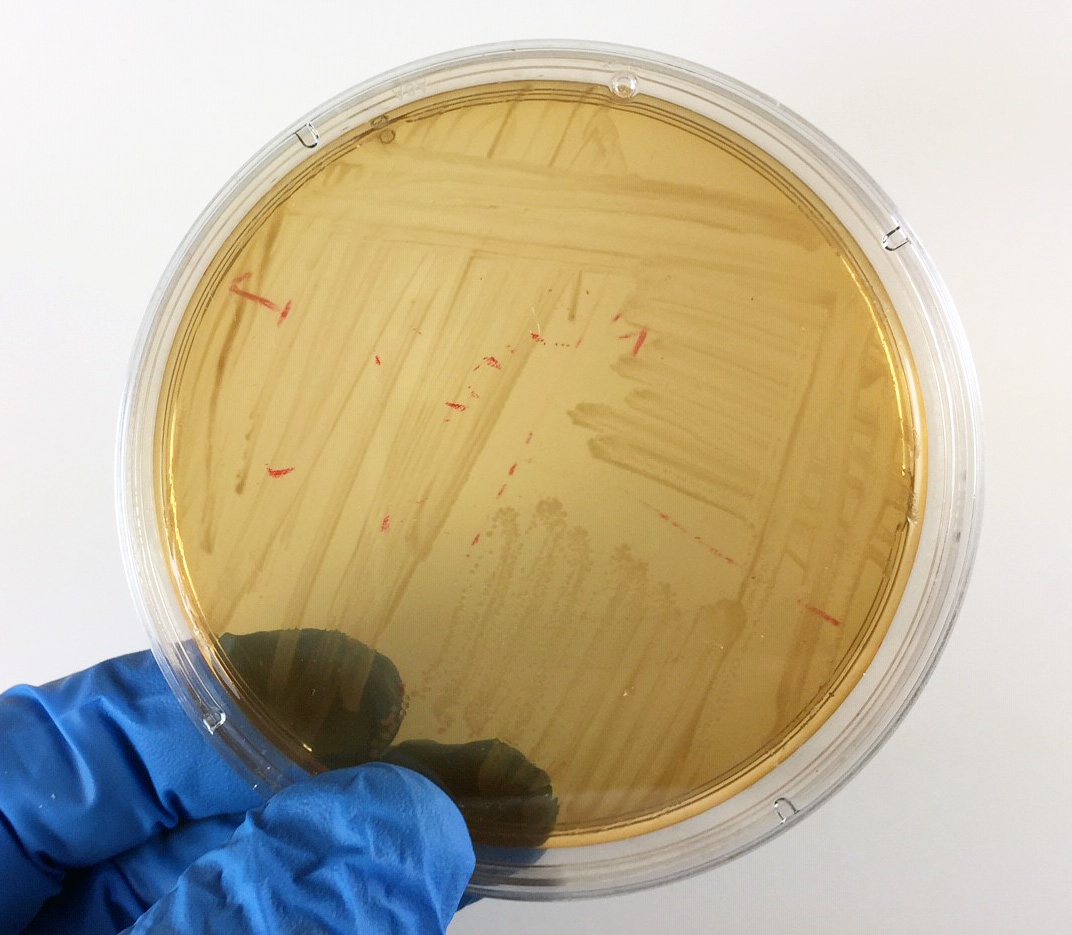
Fig. 18: Shigella Growing on XLD Agar
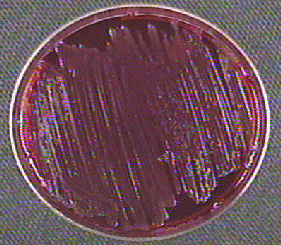
Neither lactose nor sucrose are fermented so no acid is produced. The amino acid lysine is not broken down (decarboxylated). No hydrogen sulfide is produced.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
- Sometimes the bacterium ferments the sugars producing acid end products and breaks down lysine producing alkaline end products. In this case some of the colonies and part of the agar turns yellow and some of the colonies and part of the agar turns a deeper red (see Fig. 19).
Fig. 19: Enterobacter cloacae Growing on XLD Agar
Acid from fermentation of lactose and/or sucrose lowers the pH and turns the phenol red from red (alkaline) to yellow (acid). In addition, breakdown of the amino acid lysine produces alkaline end products which raises the pH and causes some of the agar to turn a deeper red. No hydrogen sulfide is produced. 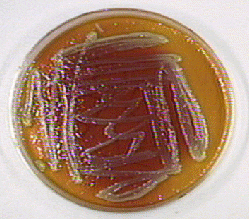
By Nathan Reading from Halesowen, UK (Salmonella species on X.L.D. agar.) [CC BY 2.0 (http://creativecommons.org/licenses/by/2.0)], via Wikimedia Commons
- If hydrogen sulfide is produced by the bacterium as a result of thiosulfate reduction, part or all of the colony will appear black (see Fig. 20). Well-isolated colonies are usually needed for good results.
Fig. 20: Salmonella Growing on XLD Agar
Neither lactose nor sucrose are fermented so no acid is produced. The amino acid lysine is broken down (decarboxylated) producing alkaline end products which cause the phenol red in the agar to turn a deeper red. Hydrogen sulfide production causes some of the colonies to appear black.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Typical colony morphology on XLD agar is as follows:
1. Escherichia coli: flat yellow colonies; some strains may be inhibited.
2. Enterobacter and Klebsiella: mucoid yellow colonies.
3. Proteus: red to yellow colonies; may have black centers.
4. Salmonella: usually red colonies with black centers.
5. Shigella, Serratia, and Pseudomonas: red colonies without black centersKeep in mind, however, that some species and subspecies do not show typical reactions.
3. Cetrimide Agar (Pseudomonas P agar)
Videos review: Cetrimide Agar Cetrimide agar contains the chemical cetrimide (cetyl timethylammonium bromide) for the selective inhibition of most bacteria other than Pseudomonas. The medium also stimulates Pseudomonas aeruginosa to produce a number of pigmented compounds, including pyoverdin and pyocyanin. The green water soluble pigment characteristic of Pseudomonas aeruginosa is due to production of a green to blue water-soluble toxin called pyocyanin (see Fig. 21). A fluorescent siderophore called pyoverdin, often produced by Pseudomonas, will typically fluoresce when the plate is placed under a short wavelength ultraviolet light (see Fig. 22). After a few minutes at room temperature, the plate loses its fluorescence. The fluorescence, however, can be restored by placing the plate back at 37°C for several minutes.
Fig. 21: Pseudomonas aeruginosa Growing on Cetrimide Agar with a Positive Oxidase Test
Fig. 22: Pseudomonas aeruginosa Growing on Cetrimide Agar and Fluorescing Under Ultraviolet Light
Note the production of pyocyanin, a green water soluble pigment, and the positive oxidase test. 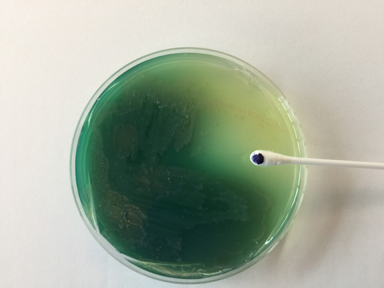
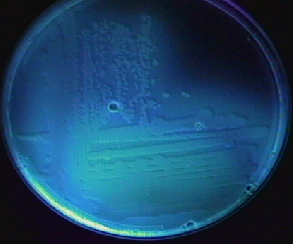
Note the production of fluorescein, a compound that fluoresces under short wavelength ultraviolet light.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
4. Oxidase Test
Videos review: Performing an Oxidase Test: Gibson Oxidase Swab Method In this lab a Gibson Oxidase Swab is used to perform the oxidase test. The oxidase test is based on the bacterial production of an oxidase enzyme. Cytochrome oxidase, in the presence of oxygen, oxidizes the N-N-N-tetra-methyl-p-phenylenediamine oxidase test reagent in a Gibson oxidase swab.
To perform the oxidase test, touch the oxidase test swab to a colony on a culture medium. An oxidase-positive reaction will appear purple to black within 10 seconds (see Fig. 8A) . An oxidase-negative test will result in no color change (see Fig. 8B). Ignore any color development occurring after 60 seconds.
Fig. 8A: A Positive Oxidase Test
Fig. 8B: A Negative Oxidase Test
Touch the Gibson Oxidase Test swab to a colony. An oxidase-positive reactions will appear purple to black within 10 seconds as shown in the above image. Disregard any color development that appears after 60 seconds.
Touch the Gibson Oxidase Test swab to a colony. An oxidase-positive reactions will appear purple to black within 10 seconds as shown in the above image. No change in color is negative. Disregard any color development that appears after 60 seconds.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Pseudomonas aeruginosa and most other glucose non-fermenting, Gram-negative bacilli are oxidase-positive. Except for the genus Plesiomonas, the glucose fermenting Enterobacteriaceae are oxidase-negative.
5. Pigment production in Pseudomonas aeruginosa
The green water-soluble pigment characteristic of Pseudomonas aeruginosa is due to production of a green to blue water-soluble toxin called pyocyanin (see Fig. 21 above). A fluorescent siderophore called pyoverdin, often produced by Pseudomonas, >will typically fluoresce when the plate is placed under a short wavelength ultraviolet light (see Fig. 22 above). After a few minutes at room temperature, the plate loses its fluorescence. The fluorescence, however, can be restored by placing the plate back at 37°C for several minutes.
6. Odor
Most of the Enterobacteriaceae have a rather foul smell; Pseudomonas aeruginosa produces a characteristic fruity or grape juice-like aroma due to production of an aromatic compound called aminoacetophenone.
7. The EnteroPluri-Test
A number of techniques can be used for the identification of specific species and subspecies of Enterobacteriaceae. Speciation is important because it provides data regarding patterns of susceptibility to antimicrobial agents and changes that occur over a period of time. It is also essential for epidemiological studies such as determination of nosocomial infections and their spread.
In an effort to simplify the speciation of the Enterobacteriaceae and reduce the amount of prepared media and incubation space needed by the clinical lab, a number of self-contained multi-test systems have been commercially marketed. Some of these multi-test systems have been combined with a computer-prepared manual to provide identification based on the overall probability of occurrence for each of the biochemical reactions. In this way, a large number of biochemical tests can economically be performed in a short period of time, and the results can be accurately interpreted with relative ease and assurance.
The EnteroPluri-Test is a self-contained, compartmented plastic tube containing 12 different agars (enabling the performance of a total of 15 standard biochemical tests) and an enclosed inoculating wire. After inoculation and incubation, the resulting combination of reactions, together with a Computer Coding and Identification System (CCIS), allows for easy identification. The various biochemical reactions of the EnteroPluri-Test and their correct interpretation are discussed below. Although it is designed to identify members of the bacterial family Enterobacteriaceae, it will sometimes also identify common biotypes of Pseudomonas and other non-fermentative Gram-negative bacilli. It does not identify Pseudomonas aeruginosa.
IDENTIFYING MEMBERS OF THE ENTEROBACTERIACEAE WITH THE ENTEROPLURI-TEST
The EnteroPluri-Test contains 12 different agars that can be used to carry out 15 standard biochemical tests (see Fig. 23). Interpret the results of your EnteroPluri-Test using the instructions below and record them on the EnteroPluri-Test table on your Results page. For more detail on the 15 biochemical tests in the EnteroPluri-Test. See Table 12A.
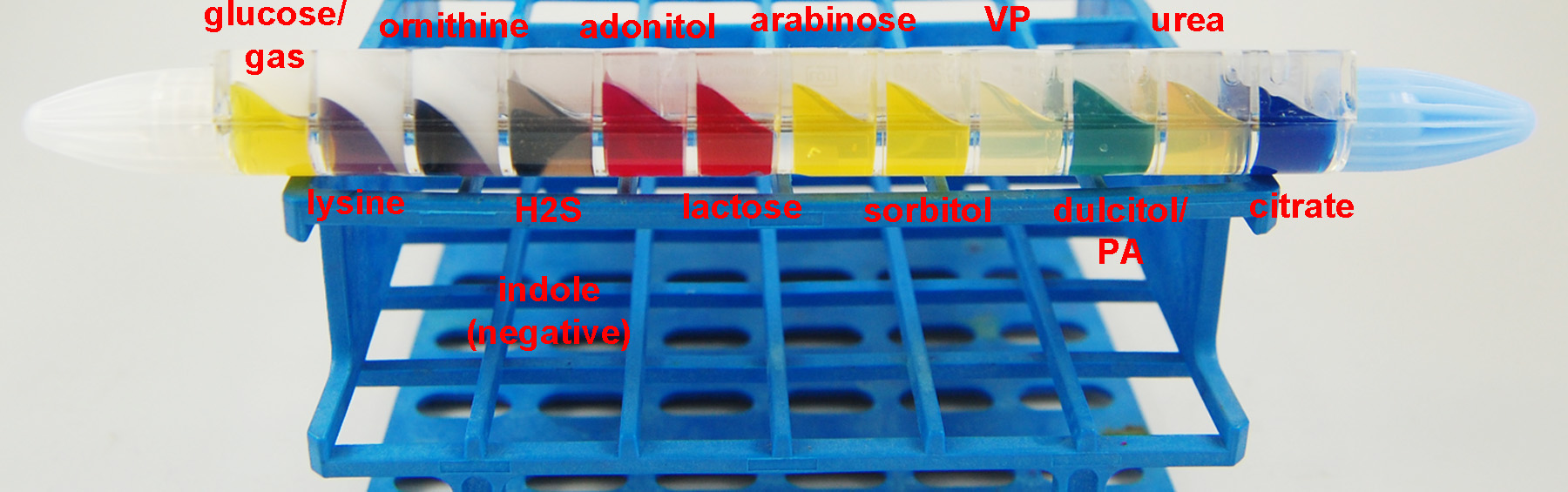
Fig. 23: The EnteroPluri-Test The EnteroPluri-Test contains 12 different agars enabling the performance of a total of 15 biochemical tests as well as an enclosed inoculating wire. Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
1. Interpret the results of glucose fermentation in compartment 1.
- Any yellow = +; red = -
- If positive, circle the number 4 under glucose on your Results page.
2. Interpret the results of gas production also in compartment 1.
- White wax lifted from the yellow agar = +; wax not lifted from agar = -
- If positive, circle the number 2 under gas on your Results page.
3. Interpret the results of lysine decarboxylase in compartment 2.
- Any violet = +; yellow = -
- If positive, circle the number 1 under lysine on your Results page.
4. Interpret the results of ornithine decarboxylase in compartment 3.
- Any violet = +; yellow = -
- If positive, circle the number 4 under ornithine on your Results page.
5. Interpret the results of H2S production in compartment 4.
- Black/brown = +; beige = - (The black may fade or revert back to negative if the EnteroPluri-Test is read after 24 hours of incubation.)
- If positive, circle the number 2 under H2S on your Results page.
6. Indole production also in compartment 4. Do not interpret the indole test at this time. Add Kovac's reagent only after all other tests have been read (see step 16 below).
7. Interpret the results of adonitol fermentation in compartment 5.
- Any yellow = +; red = -
- If positive, circle the number 4 under adonitol on your Results page.
8. Interpret the results of lactose fermentation in compartment 6.
- Any yellow = +; red = -
- If positive, circle the number 2 under lactose on your Results page.
9. Interpret the results of arabinose fermentation in compartment 7.
- Any yellow = +; red = -
- If positive, circle the number 1 under arabinose on your Results page.
10. Interpret the results of sorbitol fermentation in compartment 8.
- Any yellow = +; red = -
- If positive, circle the number 4 under sorbitol on your Results page.
11. Voges-Praskauer (VP) test in compartment 9. Do not interpret the VP test at this time. Add the reagents alpha-naphtol and potassium hydroxide (KOH) only after all other tests have been read (see step 17 below).
12. Interpret the results of dulcitol fermentation in compartment 10.
- Yellow = +; green or dark brown = -
- If positive, circle the number 1 under dulcitol on your Results page.
13. Interpret the results of PA deaminase also in compartment 10.
- Dark brown= +; green or yellow= -
- If positive, circle the number 4 under PA on your Results page.
14. Interpret the results of urea hydrolysis in compartment 11.
- Pink, red or purple = +; beige = -
- If positive, circle the number 2 under urea on your Results page.
15. Interpret the results of citrate utilization in compartment 12.
- Any blue = +; green = -
- If positive, circle the number 1 under citrate on your Results page.
16. Your instructor will add 2-3 drops of Kovac's reagent to the indole test compartment.
- Pink/red = +; yellow = -
- If positive, circle the number 1 under indole on your Results page.
17. Your instructor will add 3 drops of alpha-naphthol reagent and 2 drops of potassium hydroxide (KOH) to the VP test compartment.
- Red = +; colorless = -
- If positive, circle the number 2 under VP on your Results page.
18. Add all the positive test number values in each bracketed section and enter each sum in its code box on the EnteroPluri-Test chart on your Results page.
19. The 5 digit number is the CODICE number. Look that number up in the Codebook beginning on page 2 and identify your unknown. (Should more than one organism be listed, the confirmatory tests indicated in the CCIS would normally then have to be performed. In addition, an identification of Salmonella or Shigella would usually be confirmed by direct serologic testing as will be described in Lab 17.)
If there are any problems, consult your instructor.
Group 1
Group 2
Group 3
Group 4
Group 5
Test
Glucose
Gas
Lysine
Ornithine
H2S
Indole
Adonitol
Lactose
Arabinose
Sorbitol
VP
Dulcitol
PA
Urea
Citrate
Value
4
2
1
4
2
1
4
2
1
4
2
1
4
2
1
Result
Code
CODICE NUMBER:
8. The Complete Blood Count (CBC) Test
9. Urinalysis (The Dipstick Tests)10. SIRS and Sepsis
PERFORMANCE OBJECTIVES FOR LAB 12
After completing this lab, the student will be able to perform the following objectives:
A. ENTEROBACTERIACEAE: GLUCOSE FERMENTING, GRAM-NEGATIVE, ENTERIC BACILLI
1. Name the bacterial family to which the most commonly encountered organisms isolated from clinical specimens belong.
2. List five characteristics used to place bacteria into the family Enterobacteriaceae.
3. State what infections are caused by Salmonella and by Shigella and how they are transmitted to humans.
4. Name four strains of Escherichia coli that may infect the gastrointestinal tract.
5. Name five genera of Enterobacteriaceae considered as common opportunistic pathogens, state their normal habitat, and list four common types of opportunistic infections that they all may cause.
6. Name several predisposing factors that make one more susceptible to urinary tract infections.
7. In terms of CFUs, state the laboratory culture standards for a urinary tract infection.
8. Define nosocomial infection.
9. State the significance of endotoxins in infections caused by many of the Enterobacteriaceae.
10. Discuss the significance of R-plasmids in our attempts to treat infections caused by the Enterobacteriaceae.
B. PSEUDOMONAS AND OTHER GLUCOSE NON-FERMENTING, GRAM-NEGATIVE BACILLI
1. Name the most common glucose non-fermenting Gram-negative rod that infect humans and list five types of opportunistic infections it may cause.
2, State 3 infections being caused with increased frequency by Acinetobacter. .
C. ISOLATION OF ENTEROBACTERIACEAE AND PSEUDOMONAS
1. State the usefulness of MacConkey agar and Cetrimide agar for the isolation of Enterobacteriaceae and Pseudomonas.
D. DIFFERENTIATING BETWEEN THE ENTEROBACTERIACEAE AND PSEUDOMONAS
1. State how to differentiate Pseudomonas aeruginosa from the Enterobacteriaceae using the following tests:
a. oxidase test
b. production of pigment and fluorescent products
c. odor
E. IDENTIFYING THE ENTEROBACTERIACEAE USING THE ENTEROPLURI-TEST
Return to Menu for Lab 121. Briefly describe the EnteroPluri-Test .
SELF-QUIZ
Microbiology Laboratory Manual by Gary E. Kaiser, PhD, Professor of Microbiology
is licensed under a Creative Commons Attribution 4.0 International License.
Last updated: April, 2023