A. Staphylococcus aureus (coagulase-positive staphylococci)
Staphylococcus aureus
is the most pathogenic species and is implicated in a variety of infections.
S. aureus is with some frequency found as normal human flora in the anterior nares (nostrils). It can also be found in the throat, axillae, and the inguinal and perineal areas. Approximately 30% of adults and most children are healthy periodic nasopharyngeal
carriers of S. aureus. Around 15% of healthy adults are persistent
nasopharyngeal carriers. The colonization rates among health care workers, patients on dialysis, and people with diabetes are higher than in the general population.
In the majority of S. aureus infections the source of the organism is either:
- A healthy nasal carrier,
or
- Contact with an abscess from an infected individual.
The portal
of entry is usually the skin. S. aureus causes pus-filled
inflammatory lesions known as abscesses. Depending on the location
and extent of tissue involvement, the abscess may be called:
1.Pustules
A pustule is an infected
hair follicle where the base of the hair follicle appears red and
raised with an accumulation of pus just under the epidermis. Infected hair
follicles are also referred to as folliculitis.
2. Furuncles or boils
Furuncles appear as large, raised, pus-filled, painful nodules
having an accumulation of dead, necrotic tissue at the base. The bacteria
spread from the hair follicle to adjacent subcutaneous tissue.
3. Carbuncles
Carbuncles occur when furuncles coalesce and spread into surrounding
subcutaneous and deeper connective tissue. Superficial
skin perforates, sloughs off, and discharges pus.
S. aureus also causes
impetigo, a superficial blister-like infection
of the skin usually occurring on the face and limbs and seen
mostly in young children. S. aureus may also cause cellulitis, a diffuse inflammation of connective tissue with severe inflammation of dermal and subcutaneous layers of the skin. S. aureus is also a frequent
cause of accidental wound and postoperative wound infections.
Less commonly, S. aureus
may escape from the local lesion and spread through the blood to other body
areas, causing a variety of systemic infections that may involve every
system and organ. Such systemic infections include septicemia, septic arthritis,
endocarditis, meningitis, and osteomyelitis, as well as abscesses in
the lungs, spleen, liver, and kidneys. S. aureus pneumonia
may also be a secondary respiratory complication of viral infections such
as measles, and influenza and is a frequent cause of nosocomial pneumonia in patients who are debilitated. Finally, S. aureus is frequently introduced
into food by way of abscesses or the nasal cavity of food handlers. If it
is allowed to grow and produces an enterotoxin, it can cause staphylococcal
food poisoning.
In a 1990-1992 National Nosocomial Infections survey, CDC found S. aureus to be the most common cause of nosocomial pneumonia and operative wound infections, as well as the second most common cause of nosocomial bloodstream infections. Antibiotic resistant S. aureus is a common problem. For example, a survey conducted by CDC reported the proportion of methicillin-resistant isolates S. aureus (MRSA) with sensitivity only to vancomycin increased from 22.8% in 1987 to 56.2% in 1997.
Virulence factors for S. aureus
include exotoxins such as leukocidin (kills leukocytes), alpha and delta toxins
(damage tissue membranes), microcapsules (resist phagocytic engulfment and
destruction), coagulase and protein A (both help resist phagocytic engulfment).
Some strains also produce TSST-1 (toxic shock syndrome toxin-1) and
cause toxic shock syndrome, usually associated with wounds.
Approximately 25% of all S. aureus strains are toxigenic and approximately 6000 gases of toxic shock syndrome occur each year in the U.S.
Some strains also produce exfoliatin, an exotoxin that causes scalded
skin syndrome, an infection usually seen in infants and young children.
In the past 20 years, both community-associated and hospital-acquired infections with Staphylococcus aureus have increased. This infection rate has been accompanied by a rise in antibiotic-resistant strains - most significantly, methicillin-resistant S. aureus (MRSA) and, more recently, vancomycin-resistant S. aureus.
For further information on virulence
factors associated with S. aureus, see the following Softchalk lessons:
Since most S. aureus strains
produce the enzyme coagulase (see the coagulase test described below), they
are often referred to as coagulase-positive staphylococci.
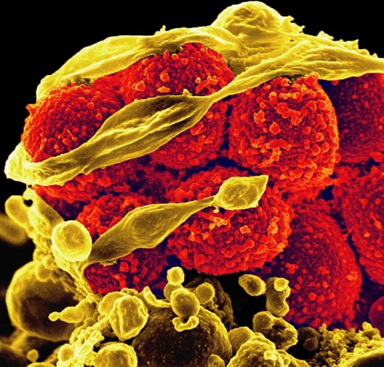
Fig. 5 shows a scanning electron micrograph of Staphylococcus aureus forming a biofilm in an indwelling catheter.
Fig. 5: Scanning Electron Micrograph Showing Biofilm Formation by Staphylococcus aureus |
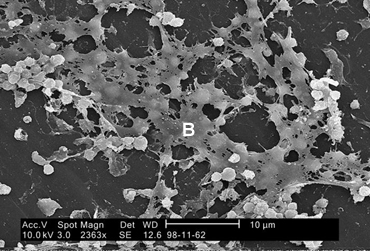 Staphylococcus aureus found on the luminal surface of an indwelling catheter. Note the polysaccharide biofilm (B) woven around the cocci.
Staphylococcus aureus found on the luminal surface of an indwelling catheter. Note the polysaccharide biofilm (B) woven around the cocci. |
By Content Providers(s): CDC/ Janice Haney Carr [Public domain]
Courtesy of the Centers for Disease Control and Prevention. |
B. Coagulase-Negative Staphylococci
Clinically common species of staphylococci
other than S. aureus are often referred to as coagulase-negative
staphylococci. These staphylococci are normal flora of the skin and, as
such, frequently act as opportunistic pathogens, especially in the
compromised host. S. saprophyticus is a relatively common cause of
urinary tract infections, especially in healthy young women, but is
seldom isolated from other sources. The great majority of infections caused
by other coagulase-negative staphylococci, including S. epidermidis,
S. haemolyticus, and S. hominis, are associated with
intravascular devices (prosthetic heart valves and intra-arterial or
intravenous lines) and shunts. Also quite common are
infections of prosthetic joints, wound infections, osteomyelitis associated
with foreign bodies, and endocarditis.
Although certain reactions may vary
from strain to strain, a series of biochemical tests will usually differentiate
the most common clinically encountered species of staphylococci. Today we will
use several tests to determine if an unknown is S. aureus, S. epidermidis,
or S. saprophyticus.
Medscape articles on infections associated with organisms mentioned in this Lab
Exercise. Registration to access this website is free.
|
ISOLATION
AND IDENTIFICATION OF STAPHYLOCOCCI
1.
Blood agar with a novobiocin (NB) disc
To isolate staphylococci, clinical
specimens are usually grown on Blood agar (described in Lab 14). Staphylococci
produce round, raised, opaque colonies 1-2mm in diameter. The novobiocin disc
is used to detect sensitivity or resistance to the antibiotic novobiocin.
| Test |
Fig. 6A: Staphylococcus
aureus (pigmented strain) on Blood Agar
Fig. 6B: Staphylococcus aureus (non-pigmented strain) on Blood Agar |
Fig. 6C: Staphylococcus
epidermidis on Blood Agar |
Fig. 6D: Staphylococcus
saprophyticuson Blood Agar |
| Hemolysis
(*) |
Usually beta(1) |
Usually gamma(2) |
Usually gamma(2) |
| Pigment |
Often creamy gold(1) |
Usually white(2) |
Usually white(2) |
| Novobiocin
test |
Sensitive |
Sensitive |
Resistant |
(*) See Lab 14 for descriptions
of hemolysis
(1) some strains do not show hemolysis and/or pigment
(2) some strains do show hemolysis and/or pigment
sensitive = zone of inhibition around disc
resistant = no zone of inhibition around disc
2.
Gram stain
All
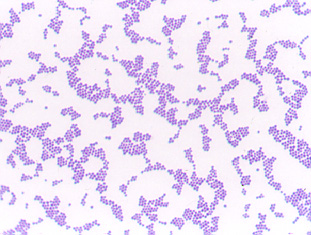
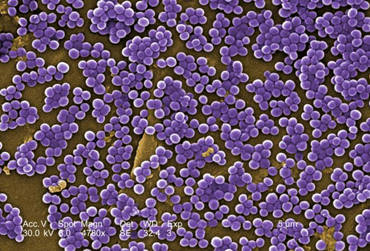
staphylococci appear as Gram-positive cocci, usually in irregular, often grape-like
clusters (see Fig. 1 above).
3.
Mannitol fermentation on Mannitol Salt agar (MSA)
Staphylococci are able to tolerate
the high salt concentration found in Mannitol Salt agar and thus grow readily.
If mannitol is fermented, the acid produced turns the phenol red pH indicator
from red (alkaline) to yellow (acid).
| Test |
Fig. 7A: Staphylococcus
aureus Growing on Mannitol Salt Agar |
Fig. 7B: Staphylococcus
epidermidis Growing on Mannitol Salt Agar |
Fig. 7C: Staphylococcus
saprophyticus Growing on Mannitol Salt Agar |
| Mannitol
fermentation |
Positive |
Negative |
Usually positive |
positive = acid end products
turn the phenol red pH indicator from red to yellow
negative = prenol red remains red
4.
Production of coagulase
The staphylococcal enzyme coagulase
will cause inoculated citrated rabbit plasma to gel or coagulate. The coagulase
converts soluble fibrinogen in the plasma into insoluble fibrin.
| Test |
Fig. 8A: Staphylococcus
aureusCoagulase Test |
Fig. 8B: Staphylococcus
epidermidisCoagulase Test |
Fig. 8C: Staphylococcus
saprophyticusCoagulase Test |
| Coagulase
production |
Positive |
Negative |
Negative |
positive = plasma will
gel or coagulate
negative = plasma will not gel
5.
The Staphyloslide® Latex Test for cell-bound coagulase (clumping factor)
and/or Protein A
The Staphyloslide® Latex Test
is an agglutination test that detects cell-bound coagulase (clumping factor)
and/or Protein A. Approximately 97% of human strains of S. aureus
possess both bound coagulase and extracellular coagulase. Approximately 95%
of human strains of S. aureus possess Protein A on their cell surface
(see Fig. 7). This test uses blue latex particles
coated with human fibrinogen and the human antibody IgG. Mixing of the latex
reagent with colonies of the suspected S. aureus having coagulase and/or
Protein A bound to their surface causes agglutination of the latex particles.
| Test |
Fig. 9A: Staphyloside® Latex Test on Staphylococcus
aureus |
Fig. 9B: Staphyloside® Latex Test on Staphylococcus
epidermidis |
Fig. 9C: Staphyloside® Latex Test on Staphylococcus
saprophyticus |
| Cell-bound
coagulase (clumping factor) and/or Protein A |
Positive |
Negative |
Negative |
positive = clumping of
latex particles
negative = no clumping of latex particles
For further information on coagulase
and Protein A associated with S. aureus, see the following CourseArc lessons:
Staphylococci are also being identified
using chemiluminescent labelled DNA probes complementary to species-specific
bacterial ribosomal RNA (rRNA) sequences as well as by other direct DNA techniques.
Return to
Menu for Lab 15
SCENERIO FOR TODAY'S LAB
Choose either unknown #1 or unknown #2 as your unknown for this Case Study.
Case Study
A 57-year- old female who is diabetic, a long-time smoker, and who 28 days ago had hip replacement surgery presents with swelling, pain, inflammation, and erythema at the surgical site. Examination shows she has a fever of 101°F, is exhibiting malaise, and has an increased total white blood cell count with a left shift. Ultrasonography examination indicates a deep abscess. A culture from an aspiration of the infected surgical site was taken.
Assume that unknown you are given is the culture from this patient.
MATERIALS
1 plate of blood agar, 1 novobiocin (NB) disc, 1 plate of mannitol salt agar, 1 tube of citrated rabbit plasma
(coagulase test), materials to perform a Gram stain, inoculating loop
PROCEDURE (to be done in groups of 3)
[Keep in mind that in a real clinical situation other lab tests and cultures for bacteria other than those upon which this lab is based would also be done.]
CAUTION: TREAT EACH UNKNOWN AS
A PATHOGEN!. Inform your instructor of any spills or accidents. WASH
AND SANITIZE YOUR HANDS WELL before leaving the lab.
|
Videos reviewing techniques used in this lab:
|
1. Do a Gram stain on the unknown (see Lab 6). Make sure you review the instructions before you do the Gram stain. Because Enterococci and Staphylococci can sometimes look similar in Gram stains done from a plate culture, perform a catalase test on your unknown to help differentiate an Enterococcus from a Staphylococcus (see Lab 8).
2. On the bottom of the petri plate, divide the plate into thirds with your wax marker and label as shown below. Before you streak your plate draw an "X" on the bottom of the blood agar plate in sector 2 to indicate where you will eventually place the Taxo NB disk as shown in Fig. 10, Step 1.
| Fig. 10:
Inoculating a Blood Agar Plate with your Unknown, Step 1 |
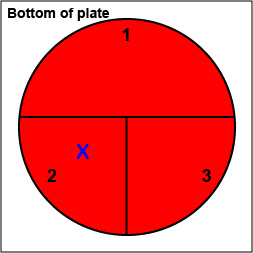 |
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
3. Using your loop, streak your
unknown for isolation on a plate of Blood agar as described
below.
a. Using a sterile inoculating loop, streak your unknown for isolation on a
blood agar plate so as to get single, isolated colonies (Fig.
10, step 2, Fig. 10, step 3, and Fig. 10, step 4).
| Fig. 10:
Inoculating a Blood Agar Plate with your Unknown, Step 2 |
Fig. 10:
Inoculating a Blood Agar Plate with your Unknown, Step 3 |
Fig. 10:
Inoculating a Blood Agar Plate with your Unknown, Step 4 |
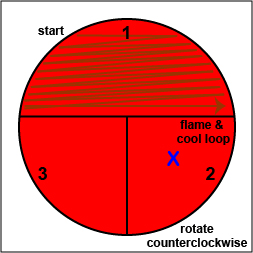 Using a sterile inoculating loop,
streak one-third of the blood agar plate with your unknown. Flame the loop and let it cool.
Using a sterile inoculating loop,
streak one-third of the blood agar plate with your unknown. Flame the loop and let it cool. |
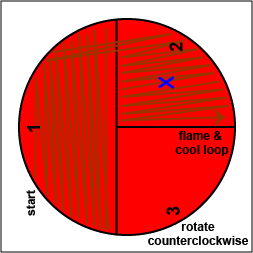 Rotate the plate counterclockwise so sector 1 is at 9:00. Using a sterile inoculating loop,
spread out some of the bacteria in area 1 over area 2. Flame the loop and let it cool.
Rotate the plate counterclockwise so sector 1 is at 9:00. Using a sterile inoculating loop,
spread out some of the bacteria in area 1 over area 2. Flame the loop and let it cool. |
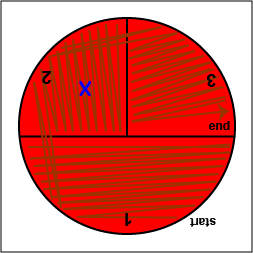 Rotate the plate counterclockwise so sector 1 is at 9:00. Using a sterile inoculating loop,
spread out some of the bacteria in area 2 over area 3.
Rotate the plate counterclockwise so sector 1 is at 9:00. Using a sterile inoculating loop,
spread out some of the bacteria in area 2 over area 3. |
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
b. Using your inoculating loop, stab the agar 2-3 times in each of the 3 growth areas in order to detect oxygen-sensitive hemolysins (Fig.
10, step 5).
| Fig. 10:
Inoculating a Blood Agar Plate with your Unknown, Step 5 |
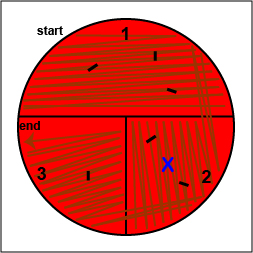 Stab the agar 2-3 times in each if the growth areas.
Stab the agar 2-3 times in each if the growth areas. |
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
c. Place a novobiocin antibiotic disk in the area where you drew the "X" in sector 2 (Fig. 10, step 6). Tap it lightly with your loop so that the disk sticks to the agar.
| Fig. 10:
Inoculating a Blood Agar Plate with your Unknown, Step 6 |
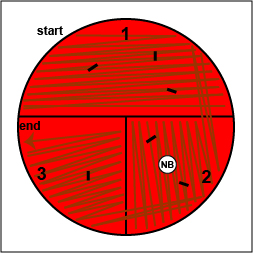 Place a Taxo NBdisk over the "X" in sector 2 and tap it lightly with your loop so that it sticks to the agar.
Place a Taxo NBdisk over the "X" in sector 2 and tap it lightly with your loop so that it sticks to the agar. |
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
d. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section until the next lab period.
4. Streak your unknown for
isolation on a plate of Mannitol Salt agar (MSA) as shown in Fig.
11. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section until the next lab period.
Fig. 1: Inoculating a mannitol salt
agar plate with Staphylococcus |
 |
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License

|
5. Inoculate a tube of citrated
rabbit plasma with your unknown and incubate your test tube rack at 37°C.
Return to
Menu for Lab 15
RESULTS
Case Study Lab Report for Lab 15:
Staphylococci
The concept behind the case studies presented in Lab 15 used to illustrate the genus Staphylococcus is for you and your lab partners as a group to:
1. Come up with a valid diagnosis of the type of infectious disease seen in your case study and identify the bacterium causing that infection.
2. Support your group’s diagnose based on:
a. Any relevant facts in the patient’s history. (A reliable on-line source will be used to support this.)
b. The patient’s signs and symptoms. (A reliable on-line source will be used to support this.)
c. Each of the individual lab tests given in your case study.
d. All microbiological lab tests you performed as part of the project.
The due date for this report can be found on the class calendar. Your grade for this lab is based on the completeness of your report and written evidence of the critical thinking process that went into making and supporting your diagnosis. Remember, you are trying to convince your instructor that you understand how the diagnosis was made by supporting that diagnosis with data.
Grading:
The lab 15 Lab Report is worth 33 points each.
These case studies are based in part on your in-class participation as part of your group. Therefore:
a. If you were not in lab when the inoculations with your unknown were performed, 3 points will be deducted from your Lab Report score for labs 12, 14, and 15; 6 points from your Lab Report score for the Final Project.
b. If you were not in lab when the results of your lab tests were observed, 3 points will be deducted from your Lab Report score for labs 12, 14, and 15; 6 points from your Lab Report score for the Final Project..
c. For each day your Lab Report is late, 2 points will be deducted from your Lab Report score for labs 12, 14, and 15; 4 points from your Lab Report score for the Final Project.
Be sure to handle all the bacterial cultures you are using in lab today as if they are pathogens! Be sure to wash and sanitize your hands well at the completion of today’s lab.
Also, make sure you observe the results of someone in your lab who had an unknown different from yours . The Performance Objectives for Lab 15 tell you what you are expected to be able to do on the practical.
Your Name:
Others in your group:
Lab section:
Date:
A. Case Study from Lab 15: Unknown #1
Case Study
A 57-year old female who is diabetic, a long-time smoker, and who 28 days ago had hip replacement surgery presents with swelling, pain, inflammation, and erythema at the surgical site. Examination shows she has a fever of 101°F, is exhibiting malaise, and has an increased total white blood cell count with a left shift. Ultrasonography examination indicates a deep abscess. A culture from an aspiration of the infected surgical site was taken.
Assume that unknown you are given is the culture from this patient.
1. Patient’s history and predisposing factors
Read the case study. Explain how any relevant parts of the patient’s history contributed to your diagnosis as to the type of infectious disease seen here. The patient's history refers to anything given in the case study prior to that patient seeking medical attention for the current medical condition. You are urged to use the computers in lab to search reliable medically oriented Internet sources to support this. Reliable sources you might consider are Medscape (http://emedicine.medscape.com/infectious_diseases)
and The Centers for Disease Control and Prevention (CDC) at http://www.cdc.gov/. Cite any sources you use at the end of this Patient's History section in APA style (https://www.scribbr.com/apa-examples/website/).
The patient's history should suggest a general type of infectious disease that is present, such as a urinary tract infection, a wound infection, gastroenteritis, pharyngitis, pneumonia, septicemia, etc. Do not look up the bacterium you eventually identify as the cause of this infectious disease. You do not know the causative bacterium at this point. You need to determine the general type of infection in order to determine what microbiological tests to perform to identify the bacterium causing the infection. Search at least one medically oriented reference article from a reliable site such as Medscape and use this article to support your diagnosis of the type of infectious disease seen here. Don't forget to cite any sources you used in APA style directly under this Patient's History and Patient's Symptoms sections of this Lab Report.
2. Patient’s signs and symptoms
Read the case study. Explain how the patient’s signs and symptoms contributed to your diagnosis as to the type of infectious disease seen here. Signs refer to anything being measured by a medical professional during a physical exam such as blood pressure, respiration rate, heart rate, oxygen saturation, and temperature. Symptoms refer to symptoms being reported by the patient. You are urged to use the computers in lab to search reliable medically oriented Internet sources to support this. Reliable sources you might consider are Medscape (http://emedicine.medscape.com/infectious_diseases)
and The Centers for Disease Control and Prevention (CDC) at http://www.cdc.gov/. Cite any sources you use at the end of this Patient's Symptoms section in APA style (https://www.scribbr.com/apa-examples/website/) Also see appendix F (SIRS and Sepsis) in your lab manual for an indication of whether or not the patient has SIRS.
The patient's signs and symptoms should suggest a general type of infectious disease that is present, such as a urinary tract infection, a wound infection, gastroenteritis, pharyngitis, pneumonia, septicemia, etc. Do not look up the bacterium you eventually identify as the cause of this infectious disease. You do not know the causative bacterium at this point. You need to determine the general type of infection in order to determine what microbiological tests to perform to identify the bacterium causing the infection. Search at least one medically oriented reference article from a reliable site such as Medscape and use this article to support your diagnosis of the type of infectious disease seen here. Don't forget to cite any sources you used in APA style directly under this Patient's History and Patient's Symptoms sections of this Lab Report.
3. Vocabulary list for medical terms used in the case study under signs and symptoms
List and define any medical terms used in your case study that describe the patients’s signs and symptoms that the average person not in the medical profession might not know.
4. Results of laboratory test given in the case study
List each lab test given in the case study that are done in a lab, such as total white blood count, differential white blood cell count, urinalysis, and X-ray, and explain how the results of that test helps to contribute to your diagnosis. The CBC test is described in Appendix C of this lab manual.
4. Microbiological lab tests you performed in Lab 15
a. Gram stain and catalase test results
Give the Gram reaction (Gram-positive or Gram negative and how you reached this conclusion) and the shape and arrangement of the unknown you were given. Because Enterococci and Staphylococci can sometimes look similar in Gram stains done from a plate culture, perform a catalase test on your unknown to help differentiate an Enterococcus from a Staphylococcus. State how this contributed to your decision as to which microbiological tests and/or media to use next. The Gram stain is discussed in Lab 6; the catalase test in Lab 8.
b. Blood agar with novobiocin (NB) disc
Give the results of the Blood agar with Taxo NB disc you performed on the unknown you were given, and how you reached this conclusion.State how this contributed to your your decision as to what bacterium is causing the infection. The possible results for Blood agar and NB disc were discussed in the beginning pages of this lab.
c. Mannitol Salt agar
Give the results of the Mannitol Salt agar you performed on the unknown you were given, and how you reached this conclusion.State how this contributed to your your decision as to what bacterium is causing the infection. The possible results for Mannitol Salt agar were discussed in the beginning pages of this lab.
d. Coagulase test
Give the results of the Coagulase test you performed on the unknown you were given, and how you reached this conclusion.State how this contributed to your your decision as to what bacterium is causing the infection. The possible results for the Coagulase test were discussed in the beginning pages of this lab.
Final Diagnosis
Genus and species of unknown #1 = ________________________________
Infection: _______________________________
B. Case Study from Lab 15: Unknown #2
Case Study
A 57-year old female who is diabetic, a long-time smoker, and who 28 days ago had hip replacement surgery presents with swelling, pain, inflammation, and erythema at the surgical site. Examination shows she has a fever of 101°F, is exhibiting malaise, and has an increased total white blood cell count with a left shift. Ultrasonography examination indicates a deep abscess. A culture from an aspiration of the infected surgical site was taken.
1. Patient’s history and predisposing factors
Read the case study. Explain how any relevant parts of the patient’s history contributed to your diagnosis as to the type of infectious disease seen here. The patient's history refers to anything given in the case study prior to that patient seeking medical attention for the current medical condition. You are urged to use the computers in lab to search reliable medically oriented Internet sources to support this. Reliable sources you might consider are Medscape (http://emedicine.medscape.com/infectious_diseases)
and The Centers for Disease Control and Prevention (CDC) at http://www.cdc.gov/. Cite any sources you use at the end of this Patient's History section in APA style (https://www.scribbr.com/apa-examples/website/). Also see appendix F (SIRS and Sepsis) in your lab manual for an indication of whether or not the patient has SIRS.
The patient's history should suggest a general type of infectious disease that is present, such as a urinary tract infection, a wound infection, gastroenteritis, pharyngitis, pneumonia, septicemia, etc. Do not look up the bacterium you eventually identify as the cause of this infectious disease. You do not know the causative bacterium at this point. You need to determine the general type of infection in order to determine what microbiological tests to perform to identify the bacterium causing the infection. Search at least one medically oriented reference article from a reliable site such as Medscape and use this article to support your diagnosis of the type of infectious disease seen here. Don't forget to cite any sources you used in APA style directly under this Patient's History and Patient's Symptoms sections of this Lab Report.
2. Patient’s signs and symptoms
Read the case study. Explain how the patient’s symptoms contributed to your diagnosis as to the type of infectious disease seen here. Signs refer to anything being measured by a medical professional during a physical exam such as blood pressure, respiration rate, heart rate, oxygen saturation, and temperature. Symptoms refer to symptoms being reported by the patient. You are urged to use the computers in lab to search reliable medically oriented Internet sources to support this. Reliable sources you might consider are Medscape (http://emedicine.medscape.com/infectious_diseases)
and The Centers for Disease Control and Prevention (CDC) at http://www.cdc.gov/. Cite any sources you use at the end of this Patient's Symptoms section in APA style (https://www.scribbr.com/apa-examples/website/).
The patient's signs and symptoms should suggest a general type of infectious disease that is present, such as a urinary tract infection, a wound infection, gastroenteritis, pharyngitis, pneumonia, septicemia, etc. Do not look up the bacterium you eventually identify as the cause of this infectious disease. You do not know the causative bacterium at this point. You need to determine the general type of infection in order to determine what microbiological tests to perform to identify the bacterium causing the infection. Search at least one medically oriented reference article from a reliable site such as Medscape and use this article to support your diagnosis of the type of infectious disease seen here. Don't forget to cite any sources you used in APA style directly under this Patient's History and Patient's Symptoms sections of this Lab Report.
3. Vocabulary list for medical terms used in the case study under signs and symptoms
List and define any medical terms used in your case study that describe the patients’s signs and symptoms that the average person not in the medical profession might not know.
4. Results of laboratory test given in the case study
List each lab test given in the case study that are done in a lab, such as total white blood count, differential white blood cell count, urinalysis, and X-ray, and explain how the results of that test helps to contribute to your diagnosis. The CBC test is described in Appendix C of this lab manual.
5. Microbiological lab tests you performed in Lab 15
a. Gram stain and catalase test
Give the Gram reaction (Gram-positive or Gram negative and how you reached this conclusion) and the shape and arrangement of the unknown you were given. Because Enterococci and Staphylococci can sometimes look similar in Gram stains done from a plate culture, perform a catalase test on your unknown to help differentiate an Enterococcus from a Staphylococcus. State how this contributed to your decision as to which microbiological tests and/or media to use next. The Gram stain is discussed in Lab 6; the catalase test in Lab 8.
b. Blood agar with novobiocin (NB) disc
Give the results of the Blood agar with Taxo NB disc you performed on the unknown you were given, and how you reached this conclusion.State how this contributed to your your decision as to what bacterium is causing the infection. The possible results for Blood agar and NB disc were discussed in the beginning pages of this lab.
c. Mannitol Salt agar
Give the results of the Mannitol Salt agar you performed on the unknown you were given, and how you reached this conclusion.State how this contributed to your your decision as to what bacterium is causing the infection. The possible results for Mannitol Salt agar were discussed in the beginning pages of this lab.
d. Coagulase test
Give the results of the Coagulase test you performed on the unknown you were given, and how you reached this conclusion.State how this contributed to your your decision as to what bacterium is causing the infection. The possible results for the Coagulase test were discussed in the beginning pages of this lab.
Final Diagnosis
Genus and species of unknown #2 = ________________________________
Infection: _________________________________
Return to
Menu for Lab 15
PERFORMANCE
OBJECTIVES FOR LAB 15
After completing this lab, the student
will be able to perform the following objectives:
DISCUSSION
1. Name three common clinically
important species of Staphylococcus and state which species is most
pathogenic.
2. State the sources
and the portal of entry for most Staphylococcus aureus infections.
3. Name and describe
three types of abscesses caused by Staphylococcus aureus.
4. Name four systemic Staphylococcus aureus infections.
5. State the significance
of Staphylococcus aureus enterotoxin, the exotoxin TSST-1, and the
exotoxin exfoliatin.
6. Name the infection
normally caused by Staphylococcus saprophyticus.
7. Name the types
of infections most commonly caused by coagulase-negative staphylococci other
than Staphylococcus saprophyticus.
ISOLATION AND IDENTIFICATION
OF STAPHYLOCOCCI
1. State the Gram reaction and
morphology of all staphylococci.
2. Describe the typical reactions
of S. aureus, S. epidermidis, and S. saprophyticus on each of
the following media:
a. Blood agar (pigment, hemolysis,
novobiocin resistance)
b. Mannitol Salt agar (for mannitol fermentation)
c. coagulase test with citrated
rabbit plasma
d. Staphyloslide® test for
bound coagulase and/or Protein A
RESULTS
1. Recognize staphylococci in a
Gram stain preparation.
2. Recognize an organism as Staphylococcus
aureus and state the reasons why after seeing the results of the following:
a. a Blood agar plate with a
novobiocin disc
b. a Mannitol Salt agar plate
c. a tube of citrated rabbit
plasma
Return to
Menu for Lab 15
SELF-QUIZ
Self-quiz
Answers
Return to
Menu for Lab 15
Lab
Manual Table of Contents

Microbiology Laboratory Manual by Gary E. Kaiser, PhD, Professor of Microbiology
is licensed under a Creative Commons Attribution 4.0 International License.
Last updated: May, 2023